+ Open data
Open data
- Basic information
Basic information
| Entry | 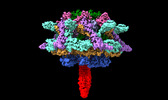 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cyanophage A-1(L) baseplate-initiators | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  baseplate / baseplate /  virus / virus /  viral protein viral protein | |||||||||
| Biological species |  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.44 Å cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Yu RC / Li Q / Zhou CZ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of the intact tail machine of Anabaena myophage A-1(L). Authors: Rong-Cheng Yu / Feng Yang / Hong-Yan Zhang / Pu Hou / Kang Du / Jie Zhu / Ning Cui / Xudong Xu / Yuxing Chen / Qiong Li / Cong-Zhao Zhou /  Abstract: The Myoviridae cyanophage A-1(L) specifically infects the model cyanobacteria Anabaena sp. PCC 7120. Following our recent report on the capsid structure of A-1(L), here we present the high-resolution ...The Myoviridae cyanophage A-1(L) specifically infects the model cyanobacteria Anabaena sp. PCC 7120. Following our recent report on the capsid structure of A-1(L), here we present the high-resolution cryo-EM structure of its intact tail machine including the neck, tail and attached fibers. Besides the dodecameric portal, the neck contains a canonical hexamer connected to a unique pentadecamer that anchors five extended bead-chain-like neck fibers. The 1045-Å-long contractile tail is composed of a helical bundle of tape measure proteins surrounded by a layer of tube proteins and a layer of sheath proteins, ended with a five-component baseplate. The six long and six short tail fibers are folded back pairwise, each with one end anchoring to the baseplate and the distal end pointing to the capsid. Structural analysis combined with biochemical assays further enable us to identify the dual hydrolytic activities of the baseplate hub, in addition to two host receptor binding domains in the tail fibers. Moreover, the structure of the intact A-1(L) also helps us to reannotate its genome. These findings will facilitate the application of A-1(L) as a chassis cyanophage in synthetic biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37151.map.gz emd_37151.map.gz | 318.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37151-v30.xml emd-37151-v30.xml emd-37151.xml emd-37151.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37151.png emd_37151.png | 57.5 KB | ||
| Filedesc metadata |  emd-37151.cif.gz emd-37151.cif.gz | 7.3 KB | ||
| Others |  emd_37151_half_map_1.map.gz emd_37151_half_map_1.map.gz emd_37151_half_map_2.map.gz emd_37151_half_map_2.map.gz | 273.7 MB 273.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37151 http://ftp.pdbj.org/pub/emdb/structures/EMD-37151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37151 | HTTPS FTP |
-Related structure data
| Related structure data |  8keaMC  8ke9C  8kecC  8keeC  8kefC  8kegC  8ts6C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37151.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37151.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37151_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
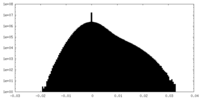
| Projections & Slices |
| ||||||||||||
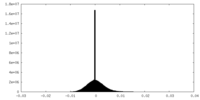
| Density Histograms |
-Half map: #1
| File | emd_37151_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
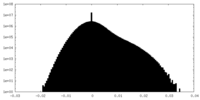
| Projections & Slices |
| ||||||||||||
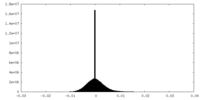
| Density Histograms |
- Sample components
Sample components
-Entire : unclassified Caudoviricetes
| Entire | Name:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
|---|---|
| Components |
|
-Supramolecule #1: unclassified Caudoviricetes
| Supramolecule | Name: unclassified Caudoviricetes / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2788787 / Sci species name: unclassified Caudoviricetes / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Nostoc sp. PCC 7120 = FACHB-418 (bacteria) Nostoc sp. PCC 7120 = FACHB-418 (bacteria) |
-Macromolecule #1: hub
| Macromolecule | Name: hub / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 98.390594 KDa |
| Sequence | String: MQDPIIKILI GNDTFLLGQE IIDLDFQIGE GKKQNNINFT IFDKDGFFAD KYISTSYAQG GIDLPIDFLE NPDDAKDTNS ASTATEVDS VGNGTVSRSG VFTPKIRAFL DTIASKETAP GTALNIEGYR SVSGSSTLFD ESEMVAGGFP RSQGSKNIGR Y QFTVIDYN ...String: MQDPIIKILI GNDTFLLGQE IIDLDFQIGE GKKQNNINFT IFDKDGFFAD KYISTSYAQG GIDLPIDFLE NPDDAKDTNS ASTATEVDS VGNGTVSRSG VFTPKIRAFL DTIASKETAP GTALNIEGYR SVSGSSTLFD ESEMVAGGFP RSQGSKNIGR Y QFTVIDYN HARSKYPNIN NYSPQNQDLL AYFKLQHRNV LPYLLRDDLD NAIDKASYEW ASFPGIGKPQ GQFNQVQSGT TI ASLKSYY ETRLAYYRSL EAGSDFQASA PKDTTNNQYA GKEYKTIRTL SNSTTASFYG YNDGFDSSDL TANGERFNPE GIT AAHESL PFGTLVKVTW AVNNKSVVVR INDRGAFVRL GRQIDLSYGA AKALSSPGND AIAAGLLTVK LEVVELRTPI GENL KESAK NQIAENLEKI KKNQKELATP EISAKGTQIT LEVSIDRSAI AVFSFLHTGT KHNAIISDTT TFTGQSVNWV LNRRV KNTR YTGVTLKGLA ATITRQYGLD LDMSEEGEMI ENISQVSQTD WQFLEKMTAI QGFGMRTVGK VLQIYKITVN AKKLNY TIS VVDNVKSLIV TDQAQTDATG SSQKIEHYGG RMTTVVDADS GSLIKVDKDN KREAGSAART FTTGVDVPQP QIQKQYS NP RPEGASVKEF QLQLELHTSQ SDLENLTPDT ALYIENTLPF IVGKSWFIES VRHSFSEGIF TSQVSAYIPV APKAIATD S GEQNSVVFDN SQSASQGVGK LPVYEILSYR SGRTVKKITS LQQSYEHWRG TGGYTAYKQL SGFSSPVSYM KGRPNQLVY DFILQQNGNQ SCPVPSPASG RVVATGGSNG MVKIDTGGGE VRLLHMSNIR VKPGQNVIRG TILGTQASVG GTSTGTHLHI EANQFILES YVQSLVTGNW |
-Macromolecule #2: central spike
| Macromolecule | Name: central spike / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 29.607072 KDa |
| Sequence | String: MIVDEFSSVY ECLLYCYKMV KRSEQLNGRR VFPILSVVTN NNDPEGRRRV KIADPLFGNL IESNWIRPIR VSQNQDNPLP QINQMVIVW FVDGDSEKGY YLPIINDANP SREKDDPVND SAVRIEGNNT IRIDKNDSET VGGNQTVAIA GEQNINVDGN L IENIGGDI ...String: MIVDEFSSVY ECLLYCYKMV KRSEQLNGRR VFPILSVVTN NNDPEGRRRV KIADPLFGNL IESNWIRPIR VSQNQDNPLP QINQMVIVW FVDGDSEKGY YLPIINDANP SREKDDPVND SAVRIEGNNT IRIDKNDSET VGGNQTVAIA GEQNINVDGN L IENIGGDI DQNVTGKIEV RSESTILIDA DGTIIIKNDS GAFISLGGNG EVLIQDSQGR KIRLGGAFNS TWDLNGLPMA FI NATSVTI AGKQIATVGA VDTRGDTIVN KGW |
-Macromolecule #3: tmp
| Macromolecule | Name: tmp / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 73.969227 KDa |
| Sequence | String: MANRQHVISV DLRLLTGNFD SSLNYIQGRL NGVFGVVSDN ATNSFFGNGG FLSSYLWNQA FNAIQMAGIA AFRAIGNEVK RAISYQDNL ISMATSYKGL LNLTTKEAFG LARESNIIFQ QRNVGLPTFG NFGTQLRAQY GDDFLSAFYD QNNMKGSLER T ADMIGRTT ...String: MANRQHVISV DLRLLTGNFD SSLNYIQGRL NGVFGVVSDN ATNSFFGNGG FLSSYLWNQA FNAIQMAGIA AFRAIGNEVK RAISYQDNL ISMATSYKGL LNLTTKEAFG LARESNIIFQ QRNVGLPTFG NFGTQLRAQY GDDFLSAFYD QNNMKGSLER T ADMIGRTT VLLGSISGVT NYQRTSFLSN FLSDDFKKLQ RLEVFRNSPQ LQRAFNSQVD SIGGEQGFNA LTRKDRLLLL EN IAAIAVD DDIIKAYKDS LGGVLSRLGQ ELFGDIGIFS FTRDLIFGLP NTSILNSITF FMKEVFAPDG VFGLIKPALQ GIV DTVLIG TKFILDRTSF IFMGLNALAR VLQPIAPLIY ATSSALTVLA AVGLANTIKS AVQGYLISSG AAMLGGRMGM AAMF GMGGD LVGLKTLGTL GLGKLIAPLG FLTMGLDKLA DVGWLAISGG FAKAALGIKT FGMSLWAIVA NPAFLTIAGI VGAIA LAGY TLWKYWEPIG DFLQGFWIGF KAGLPILDQI ESVLQSIGQA LSPITEGFKN MASAMQPVLD GFTKFFNLQQ EKGGEN YVK QGVNFGDAAS GVTRTILSNP LINPFSGFFG LFGNPAQNKA DGDPQSVMSA INDERRKMPS GASLVIANSS ENVFTRS QT QQLASAGRGS TVINNTFNIS GADPQAIAKQ IADIINSQWQ GTLTDYA |
-Macromolecule #4: baseplate gp16
| Macromolecule | Name: baseplate gp16 / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 17.151273 KDa |
| Sequence | String: MTTQEFLSKN KTIESELLDL LIKPNTDDSI LTRNKQAIAD RDLFDIEWEP GQSLNKLATE YLGDSFAWQI IADANGIDPT KEIDIGAGL KVPDQKALEN SIKKFIVNSP TGKQLISDAK QSILNLIGVG DSNTEFSKTL KDCIGKVVNF SFDNTQ |
-Macromolecule #5: tube initiator
| Macromolecule | Name: tube initiator / type: protein_or_peptide / ID: 5 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 31.966795 KDa |
| Sequence | String: MSKFLKNRAV VNGLPVIKNP GKYQHCYLIE YEDSTNVKQT PTENKNKQQQ GFPVYLFMMN PENITYNLPI NYQEIAIPFT AKNQLNYSN GGNIVMTMSN LILDTMDEKR SLQPLIDRLI ALREPTVKKG LKSHPKILAF KWGSNTFAPC VLTNISFDVT R WIDGYPTK ...String: MSKFLKNRAV VNGLPVIKNP GKYQHCYLIE YEDSTNVKQT PTENKNKQQQ GFPVYLFMMN PENITYNLPI NYQEIAIPFT AKNQLNYSN GGNIVMTMSN LILDTMDEKR SLQPLIDRLI ALREPTVKKG LKSHPKILAF KWGSNTFAPC VLTNISFDVT R WIDGYPTK ARVNMSLKEI QKPSSDSKAL EEAKKKVKVE TVQNGNLKKT LSEKQLIDGV KRVTEYLKKN ISFQPRTIQN IL SDPKSVI KIDKDTGQVS LFNGNGEFAA LVGTYNGDIF SPSRQ |
-Macromolecule #6: sheath initiator
| Macromolecule | Name: sheath initiator / type: protein_or_peptide / ID: 6 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 12.997665 KDa |
| Sequence | String: MYVYPPRIIN GTLYKSTDLA EYTRGKILQT LSIVRGELVA DPFFGVPLRL FSNINDYQSD VSKIQIILEE EITEAQFIVT GDLKNNGIV SLTVYWAYLG TENIENFEID TSNIF |
-Macromolecule #7: wedge protein gp31
| Macromolecule | Name: wedge protein gp31 / type: protein_or_peptide / ID: 7 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 42.385879 KDa |
| Sequence | String: MTTDLNAPQL VVDDYEQLII DSLVHTNVVS NGEFTDLDAS GFMRPFAGTM AYAGSELLYK ANLASIAAAK SFFKNVLGVP EDTGTKATT TLQFGLSASL STDFIVPINF QVSDLSGTLR FYTIGNLVIP AGATFGTIEA IAEDIGEKYN VSANFIDQYS T PLTYLQYV ...String: MTTDLNAPQL VVDDYEQLII DSLVHTNVVS NGEFTDLDAS GFMRPFAGTM AYAGSELLYK ANLASIAAAK SFFKNVLGVP EDTGTKATT TLQFGLSASL STDFIVPINF QVSDLSGTLR FYTIGNLVIP AGATFGTIEA IAEDIGEKYN VSANFIDQYS T PLTYLQYV TNIRPATNGR SGETIDNLIE RCAQIIRIRN PVSALDFEQL AELTMGEGSR CKAIGLLGIN KIVTDPQPGV VH LFLLDVN GNPADPVTIS TVGATLQPRI MLGTRLLISP MEVLNIELEL IALSDSSKTF QQLADDILEA LKVFFNPANL TPG EPVLIE EVKFAIRSVG GLSISYLQMN DNAINIPMPN QWTIPRFSYI GFELTDSEGT VYRDNVVTVT NPEE |
-Macromolecule #8: wedge protein gp32
| Macromolecule | Name: wedge protein gp32 / type: protein_or_peptide / ID: 8 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 22.423377 KDa |
| Sequence | String: MSKDAFTAWN VDRRPIYSRM PKEQVGTSYH DYIATDWLTA YWDKIFIECY DKLEDLPRQF DPLQCDEEYL DFLAPLCGWT APYWSGDYP PESKRVLLAN SYSLIWRDKG SLTVLSFVLN ALFINHRIFV PGSFILGQSQ VSEDTLGAAG WEFEILLPRD Y AENGYEFR LTLKIASLFS PLWCKYRVRY DNL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 300 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.44 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 41062 |
 Movie
Movie Controller
Controller



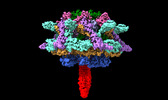
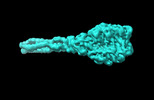
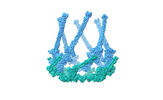



 Z
Z Y
Y X
X