+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
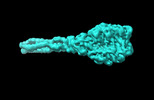
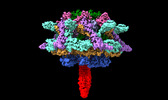
| Title | Cyanophage A-1(L) sheath-tube | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  tail / tail /  virus / virus /  viral protein viral protein | |||||||||
| Biological species |  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.26 Å cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Yu RC / Li Q / Zhou CZ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of the intact tail machine of Anabaena myophage A-1(L). Authors: Rong-Cheng Yu / Feng Yang / Hong-Yan Zhang / Pu Hou / Kang Du / Jie Zhu / Ning Cui / Xudong Xu / Yuxing Chen / Qiong Li / Cong-Zhao Zhou /  Abstract: The Myoviridae cyanophage A-1(L) specifically infects the model cyanobacteria Anabaena sp. PCC 7120. Following our recent report on the capsid structure of A-1(L), here we present the high-resolution ...The Myoviridae cyanophage A-1(L) specifically infects the model cyanobacteria Anabaena sp. PCC 7120. Following our recent report on the capsid structure of A-1(L), here we present the high-resolution cryo-EM structure of its intact tail machine including the neck, tail and attached fibers. Besides the dodecameric portal, the neck contains a canonical hexamer connected to a unique pentadecamer that anchors five extended bead-chain-like neck fibers. The 1045-Å-long contractile tail is composed of a helical bundle of tape measure proteins surrounded by a layer of tube proteins and a layer of sheath proteins, ended with a five-component baseplate. The six long and six short tail fibers are folded back pairwise, each with one end anchoring to the baseplate and the distal end pointing to the capsid. Structural analysis combined with biochemical assays further enable us to identify the dual hydrolytic activities of the baseplate hub, in addition to two host receptor binding domains in the tail fibers. Moreover, the structure of the intact A-1(L) also helps us to reannotate its genome. These findings will facilitate the application of A-1(L) as a chassis cyanophage in synthetic biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37153.map.gz emd_37153.map.gz | 316.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37153-v30.xml emd-37153-v30.xml emd-37153.xml emd-37153.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37153.png emd_37153.png | 52.8 KB | ||
| Filedesc metadata |  emd-37153.cif.gz emd-37153.cif.gz | 5.4 KB | ||
| Others |  emd_37153_half_map_1.map.gz emd_37153_half_map_1.map.gz emd_37153_half_map_2.map.gz emd_37153_half_map_2.map.gz | 273.2 MB 272.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37153 http://ftp.pdbj.org/pub/emdb/structures/EMD-37153 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37153 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37153 | HTTPS FTP |
-Related structure data
| Related structure data |  8keeMC  8ke9C  8keaC  8kecC  8kefC  8kegC  8ts6C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37153.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37153.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37153_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37153_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : unclassified Caudoviricetes
| Entire | Name:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
|---|---|
| Components |
|
-Supramolecule #1: unclassified Caudoviricetes
| Supramolecule | Name: unclassified Caudoviricetes / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2788787 / Sci species name: unclassified Caudoviricetes / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Nostoc sp. PCC 7120 = FACHB-418 (bacteria) Nostoc sp. PCC 7120 = FACHB-418 (bacteria) |
-Macromolecule #1: sheath
| Macromolecule | Name: sheath / type: protein_or_peptide / ID: 1 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 54.04541 KDa |
| Sequence | String: MTNFLNGVNI GTPGAYAFYQ TTQSRPINVE PFRTCYMVGF ASNGVNKNVP TRISNLTDFT NVYGTSASTN SVDLFFKNSQ GFGNLYFVN VAIPTRYQIV VTAATAGSYS VTVNGVTKAI TVVGGATTTT IAADVISAIN NDTVLNKEVL ATVGGTSSTV V ITSKKPTN ...String: MTNFLNGVNI GTPGAYAFYQ TTQSRPINVE PFRTCYMVGF ASNGVNKNVP TRISNLTDFT NVYGTSASTN SVDLFFKNSQ GFGNLYFVN VAIPTRYQIV VTAATAGSYS VTVNGVTKAI TVVGGATTTT IAADVISAIN NDTVLNKEVL ATVGGTSSTV V ITSKKPTN TTTAAVTGVI FTLTTTTGTS PSVADYVYTI NNTFDPALEA GFVIAPEAFS TFTKSDRLSI QVALENLCSA YR YQWAALI DSGAMSEISN TDRAIAEAAT YNSVQGHCSY YYPYLINLDD QQVPPSAAVA GMALYRFVID GFAEPPAGVN FPL KGVKNV AYKVTWEEQN VANPEGVNCI LNKENYGIVV WGARTLSADP NIVFISTRII LNIVINTLNR GYDFDIFNSV GGTA TVLDN IQRKTNTLLT TLYQAGLFYG QTTSEAFSVL GDASVQVPSL LQQGLVNMFI WVVPSTIIER LIINIKQTAI GDLEA TVAL DTAALQSSVE EGTATEGTAP VV |
-Macromolecule #2: tube
| Macromolecule | Name: tube / type: protein_or_peptide / ID: 2 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  unclassified Caudoviricetes (virus) unclassified Caudoviricetes (virus) |
| Molecular weight | Theoretical: 18.55791 KDa |
| Sequence | String: MAVSKRPFSI NSFAVNLNIG NFVDARYWSK CSKIEKTYNT GEYSDGQSNI IYTLPGAIKY PEVVLSKAFS PGDEELINRL IAVNSDPIA WVTVFIQPMY RDGYYNVPQG GKIILEFCTV ARATPINEID TIGSNAAMFE CALNPSRIRS DGGNINWWSE P AAQVAEF |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 300 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.26 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 41062 |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X