+Search query
-Structure paper
| Title | Structural basis of Acinetobacter type IV pili targeting by an RNA virus. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 2746, Year 2024 |
| Publish date | Mar 29, 2024 |
 Authors Authors | Ran Meng / Zhongliang Xing / Jeng-Yih Chang / Zihao Yu / Jirapat Thongchol / Wen Xiao / Yuhang Wang / Karthik Chamakura / Zhiqi Zeng / Fengbin Wang / Ry Young / Lanying Zeng / Junjie Zhang /  |
| PubMed Abstract | Acinetobacters pose a significant threat to human health, especially those with weakened immune systems. Type IV pili of acinetobacters play crucial roles in virulence and antibiotic resistance. ...Acinetobacters pose a significant threat to human health, especially those with weakened immune systems. Type IV pili of acinetobacters play crucial roles in virulence and antibiotic resistance. Single-stranded RNA bacteriophages target the bacterial retractile pili, including type IV. Our study delves into the interaction between Acinetobacter phage AP205 and type IV pili. Using cryo-electron microscopy, we solve structures of the AP205 virion with an asymmetric dimer of maturation proteins, the native Acinetobacter type IV pili bearing a distinct post-translational pilin cleavage, and the pili-bound AP205 showing its maturation proteins adapted to pilin modifications, allowing each phage to bind to one or two pili. Leveraging these results, we develop a 20-kilodalton AP205-derived protein scaffold targeting type IV pili in situ, with potential for research and diagnostics. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:38553443 / PubMed:38553443 /  PubMed Central PubMed Central |
| Methods | EM (helical sym.) / EM (single particle) |
| Resolution | 3.0 - 8.55 Å |
| Structure data | EMDB-41442, PDB-8tob: EMDB-41443, PDB-8toc:  EMDB-41447: AP205 binding to one Acinetobacter GP16 type iv pilus EMDB-41634, PDB-8tv9: EMDB-41635, PDB-8tva: 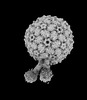 EMDB-41646: AP205 phage Acinetobacter gp16 T4P complex EMDB-41657, PDB-8tw2: EMDB-41666, PDB-8twc: |
| Source |
|
 Keywords Keywords |  CELL ADHESION / T4P / Competence / VIRUS/RNA / CELL ADHESION / T4P / Competence / VIRUS/RNA /  Acinetobacter / SsRNA phage virus / Acinetobacter / SsRNA phage virus /  VIRUS / VIRUS-RNA complex / VIRUS / VIRUS-RNA complex /  VIRUS LIKE PARTICLE / VLP VIRUS LIKE PARTICLE / VLP |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





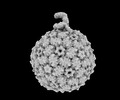








 acinetobacter genomosp. 16bj (bacteria)
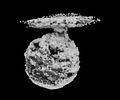
acinetobacter genomosp. 16bj (bacteria) bacteria abnormis (insect)
bacteria abnormis (insect)
