+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acinetobacter phage AP205 | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Acinetobacter / SsRNA phage virus / Acinetobacter / SsRNA phage virus /  VIRUS / VIRUS-RNA complex VIRUS / VIRUS-RNA complex | |||||||||
| Function / homology | Assembly protein / Phage maturation protein / virion attachment to host cell pilus /  virion component / virion component /  viral capsid / Coat protein / Maturation protein viral capsid / Coat protein / Maturation protein Function and homology information Function and homology information | |||||||||
| Biological species |  Bacteria abnormis (insect) / Bacteria abnormis (insect) /   Acinetobacter phage AP205 (virus) Acinetobacter phage AP205 (virus) | |||||||||
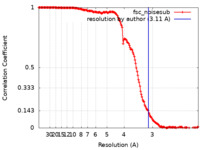
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.11 Å cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Meng R / Xing Z / Chang J / Zhang J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis of Acinetobacter type IV pili targeting by an RNA virus. Authors: Ran Meng / Zhongliang Xing / Jeng-Yih Chang / Zihao Yu / Jirapat Thongchol / Wen Xiao / Yuhang Wang / Karthik Chamakura / Zhiqi Zeng / Fengbin Wang / Ry Young / Lanying Zeng / Junjie Zhang /  Abstract: Acinetobacters pose a significant threat to human health, especially those with weakened immune systems. Type IV pili of acinetobacters play crucial roles in virulence and antibiotic resistance. ...Acinetobacters pose a significant threat to human health, especially those with weakened immune systems. Type IV pili of acinetobacters play crucial roles in virulence and antibiotic resistance. Single-stranded RNA bacteriophages target the bacterial retractile pili, including type IV. Our study delves into the interaction between Acinetobacter phage AP205 and type IV pili. Using cryo-electron microscopy, we solve structures of the AP205 virion with an asymmetric dimer of maturation proteins, the native Acinetobacter type IV pili bearing a distinct post-translational pilin cleavage, and the pili-bound AP205 showing its maturation proteins adapted to pilin modifications, allowing each phage to bind to one or two pili. Leveraging these results, we develop a 20-kilodalton AP205-derived protein scaffold targeting type IV pili in situ, with potential for research and diagnostics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41443.map.gz emd_41443.map.gz | 185.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41443-v30.xml emd-41443-v30.xml emd-41443.xml emd-41443.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41443_fsc.xml emd_41443_fsc.xml | 17.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_41443.png emd_41443.png | 60 KB | ||
| Filedesc metadata |  emd-41443.cif.gz emd-41443.cif.gz | 8.1 KB | ||
| Others |  emd_41443_half_map_1.map.gz emd_41443_half_map_1.map.gz emd_41443_half_map_2.map.gz emd_41443_half_map_2.map.gz | 344.8 MB 344.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41443 http://ftp.pdbj.org/pub/emdb/structures/EMD-41443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41443 | HTTPS FTP |
-Related structure data
| Related structure data |  8tocMC  8tobC  8tv9C  8tvaC  8tw2C  8twcC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41443.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41443.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map a
| File | emd_41443_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map a | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map b
| File | emd_41443_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map b | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Acinetobacter phage AP205
| Entire | Name:   Acinetobacter phage AP205 (virus) Acinetobacter phage AP205 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Acinetobacter phage AP205
| Supramolecule | Name: Acinetobacter phage AP205 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1, #3, #2 Details: amplified and purified from infected Acinetobacter GP16 cells. NCBI-ID: 154784 / Sci species name: Acinetobacter phage AP205 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Acinetobacter genomosp. 16BJ (bacteria) Acinetobacter genomosp. 16BJ (bacteria) |
| Molecular weight | Theoretical: 3.94 MDa |
| Virus shell | Shell ID: 1 / Name: Coat / Diameter: 290.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: RNA (4269-MER)
| Macromolecule | Name: RNA (4269-MER) / type: rna / ID: 1 / Details: genomic RNA / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Bacteria abnormis (insect) Bacteria abnormis (insect) |
| Molecular weight | Theoretical: 1.368509375 MDa |
| Sequence | String: GGAGUGAACC CCGGAGGGGG UUCGCUGAAA GCCGAAUCGA AUUCGACUUU GCGUGAUUCA CAUCACGUCU UACUCACGAU ACUAGUACC GCGAGUUAUC UUGUGGUAAU UAAAAACUAC CAGGAGAUAA CUUUAUGAAG AAAAGGACAA AAGCCUUGCU U CCCUAUGC ...String: GGAGUGAACC CCGGAGGGGG UUCGCUGAAA GCCGAAUCGA AUUCGACUUU GCGUGAUUCA CAUCACGUCU UACUCACGAU ACUAGUACC GCGAGUUAUC UUGUGGUAAU UAAAAACUAC CAGGAGAUAA CUUUAUGAAG AAAAGGACAA AAGCCUUGCU U CCCUAUGC GGUUUUCAUC AUACUCAGCU UUCAACUAAC AUUGUUGACU GCCUUGUUUA UGUAUUACCA UUAUACCUUU UA GGAGAUG GUGUCAUGAA CAUGUACAAA UGGGUACCUG AAAGUAUCCG CGAUUCUGGC GAGGGGCAAC CCUCUUAUUC AAA UAAUGG UGAUUAUGCA CCGAGCGGCC CUUGGGUUGC UGCGGGUAUU CAUACCAUGC CACAAUCGCU GCGGGAUUCC AUGA GAAAU UCUAUCAUGG UCACCGCGCA AGCUCGUCGU GAUGUCAUUG GCCCCGAAUG GGGCCCUGAC GGACGCUUUA CUGGA UAUG CUUCAGUGAU CGGGACACCU GAUCCUAAGC CUGCUGAUAU UGUGAACAAG UUUACAGUUG AACGCAGACC GGUCAG CAA CGGAAAUUUU CAACAGCGUG UGAAAGCUGG UGACAUUGUU GUUGCACCGU AUACCAGUGA UGGAAAGAUU ACUGUUA AA CUAGUCGCCG GUCAGAAGGA CAUUUCAAGU ACUCCUGAUU ACGAUUAUCG AAUUGACAGU AGUUUGGCGU CAUCCGCC G GAUUUGUUGU UGCUGGUGAA CGUUGGUAUU AUACCAAACG UCACUUCAUU AUCCCUCGUU ACUUCCAAAA CUGGCGCAU GCGCCGGCGU AAGUACGUAA CUGGUUGGGU AAUGCCAACG UUUUAUAGUC CGAAAGAGAU UUUUAAUCGC CUUAAGGAUU CGUUGGUAC CAGAUACUGG GUUAGUCACC CAAGUUUGGG CAGACAACAA CACAAAACGG AUGGAUUUCC UCACCGCUAU G GCUGAAAU CCCACAGACU CUCUCUUCUU UUCUCGAUGC GUUGGGUUAC CUCGGAUCGC UUAUUAAAGA UUUUAAACGU CG UCGCUUC UUUUUAAAUA AAGCGCAUCA ACGUAUCCGU AAUAAGCUCG GGGUGUCUUU CGCAGAAAGA AGAUCACAAA UUG UAUCUA AGUACGAUCG UAAGAUCGCA UCUGCCCGUA AGCCUGCAAU UAUUGUAAAA UUGCGGCAAC GGAAAGAAAA GGCC UUAAA AGCCCUAGAU AAAAUGCGUG UUCGAGAGGA AAAGAAAAUG AUACGUGAAU UUGCCACUCA GGCAGCCUCA CUAUG GCUU UCUUUUCGGU ACGAGAUCAU GCCGCUUUAU UAUCAAUCUC AGGACGUAUU GGACGUAAUU GCCAACUCGA CUUCUG AAU UUAUGACAUC GCGGGACUUU GUUGCUAAAG CAAUCAACAU UGGAAUUCCU UUGGAAUGGA AUCUUGAUCA AGAAAAC UU GGUUUCUCAA CCGAGACACA AUGUGAUGGU UAAAUCAAAA UUGUCACCCG AAAACAACAU CGGGAAGACU CUUUCAGU U AAUCCAUUUA CAACAGCUUG GGAGCUGUUG ACAUUGUCCU UCGUCGUCGA CUGGUUUGUC AACUUUGGUG ACGUCAUCG CAGGGUUUAC UGGCGGUUAC UCAGAUGAUU CUGGGGCAAC UGCUAGUUGG CGCUUUGAUG AUAAAAAGGU AUUCCACUUA AAGAAUAUC CCCUCAGCUA UGGUGAUCGU CGACAUUAAC UUCUACACCC GUCAGGUCAU UGACCCGCGG CUGUGCGGGG G GCUUGCUU UCUCCCCCAA ACUUAACCUU UUCCGGUAUC UUGACGCCAU GAGUUUAUCA UGGAAUCGAU CUCGUUUAAA GA UCAGUCG AGCUACUUGA CAAUUUUCUG CGCACCCAUC CCGGGUGGCG CCCAAAGUGA GGAAAAUCAC AUGGCAAAUA AGC CAAUGC AACCGAUCAC AUCUACAGCA AAUAAAAUUG UGUGGAGUGA UCCAACUCGU UUAUCAACUA CAUUUUCAGC AAGU CUGUU ACGCCAACGU GUUAAAGUUG GUAUAGCCGA ACUGAAUAAU GUUUCAGGUC AAUAUGUAUC UGUUUAUAAG CGUCC UGCA CCUAAACCGG AAGGUUGUGC AGAUGCCUGU GUCAUUAUGC CGAAUGAAAA CCAAUCCAUU CGCACAGUGA UUUCAG GGU CAGCCGAAAA CUUGGCUACC UUAAAAGCAG AAUGGGAAAC UCACAAACGU AACGUUGACA CACUCUUCGC GAGCGGC AA CGCCGGUUUG GGUUUCCUUG ACCCUACUGC GGCUAUCGUA UCGUCUGAUA CUACUGCUUA AGUGGUGAUU ACUGUGCC U AAAAGUCAAA AUAAACGACA AAUAAGACGC AGUUCUUCCG UUAAUUACAA GAAUAUCGUU AAAGCUUGCA AUGAUGCAA UGCUAAACGC UUGUGAUCAA CUGAAGUCCA CGAGUAUUCC UGCUUUCCAA UCAAACGUCC UUUCGGAUGU UCUUUCCCUC UCUGAUGCG GCCGACAUAA CAGUCAAGCA CCGAAUUGUU UCUAAAUUCG GCGAGCCUGC UGGGUCGAGC CUCCGCGACG U UGCUUUUA ACAAUUAUAA AUUGUUCGAA CAACAUCUUG GGAGCAUUCC UCAGAUUACU AAUCUGUGGC AGGAAGGAAA AG AGUUUUU CUUUUUGCGG AAAGCAAAGG CUAACUUGGG UAAAUGGUUA AAAACAUUUA AACUUGACUA UAAUUCUAUU ACA GUCGAG UUCACCCCAG GUGAGUCUUA UACCUCGGCC ACUGGGCACG UAUCGGUGUU UGCUAAGCUU UCCAACUUAG CUCA CUGGA CAUGCACUGC UGACGUCGUU GAUGAUGUUU GCCAUCUAGU GUAUUAUAAU CGCGGCCUAA AGGCUGCCGC UAGAA AACA CAUCGGUCUG AUGGUCCCAA UUGAGGGAGA GUCUGGGUUU GACACCUUUU CUCGCCACCU CAUGGGUGUU AUAUCC AUC GUUCCUGGGG CCCGCGGCGC AUCCGUGCCG AAGAACCAGG AAACGGACCG UUUUAUCGAC GUUGAACCCA CUUUCAA UA UGAUUCUCCA GCGUUGGGUA GCGGGCGAAA UUACUCGCUG CUUAACUUUA GCUAAGAAUC AUCUUGGCGC AUCACGGA A UAUUAACGGU AAAGUUGUAU UUCACGAUGC UCAAGAAUUG CACAAAGAAA UGAUCCGAGA UCUUUCUUAU GCUACUAUU GAUUUUUCAA ACGCUUCUGA UAGCGUCUUG CUGUGGGUGG UACAGCUUCU UUUUCCGAAG CAUGUAUCGU AUGUUUUGAC ACAGUAUCG UUCGUCGACU GUCCAACUCG GUUCAGAUCU UAUCGAACCG AAUAAACUUU CAAGUAUGGG AAAUGGUUUU A CUUUUGAA GUAAUGACCC UCCUCUUACU GUCGAUAGGU AGAAUCUUUG AUCCUACCUG CCGGGUUUAC GGAGAUGAUG UU AUCAUCA AAGCAGAAGU AGCCGACGAU UUCAUCAACA CUGUGUCAUC CAUUGCCUUC AUGACGAACA AUAAGAAGAC CUU UUUGAA GGGUCUCUUU CGUGAAUCAU GCGGUGCUUU CCAAUUUGAC ACAUUUGACA UCCAGUCAUU UGAGUUCGAA UGGG CUGAU AAUUUUACUG ACGUUAUUGC GAUCUGCAAC AAACUGAAGU UAAUUAUCGA CGCUGCUCAA UGCAACGAAG CAGUA AUAG CAAUAUUACG CAAUGCGCAU ACCGUCAUCU GUGAAUGCAU CCCUGUUCUU UGCAAGGGAC CGCAGCCGCC UGAUUU CAA CCUCUUUUUA UCUCAAUAUG UUUAUGAUGA UAAUUGGAAG AAGAAACAGA UGAAAUCUGA UUUAGCCAUA ACUAAGC UA AAUAGACUCG UUGAUAAACA AUGGGGUUUC UUUUCAGCUA CACAUCAUCA CCCUGAGGAA UUAUGUUACG UAAACAUU C CUGUUUACGU CCCUCGUCGU GAUUCUGUUC AUGCUGGCCA GAAUCUUUUC GUUGACCUUU CAAAUCUUUA CGCUUUACG UUUUACCAAA UCAACGGUAA GAGGUAAAGG UAAAUGGGUC AAUGUUCCCC ACUGGGUUAC ACCGGUUGGU UCAAUUUAUC GUGCUUCCC GUAUCAGACA GCAAUACCCU AACAUAGGGG AAUUGCCUAC CUGCUACUGG UCACCACAUC AGUUGGACUU G AUCACCUC CUAAUAAAUC UUUACGAUUU AUAAUAAUGG UAUGUACUAU GAGUAUGUAU GUAGGUUGAA AACCCUACCC GC UUAGGAU UGCUUAGCAG UCCUUCCCGG CA |
-Macromolecule #2: Maturation protein
| Macromolecule | Name: Maturation protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Acinetobacter phage AP205 (virus) Acinetobacter phage AP205 (virus) |
| Molecular weight | Theoretical: 61.063852 KDa |
| Sequence | String: MNMYKWVPES IRDSGEGQPS YSNNGDYAPS GPWVAAGIHT MPQSLRDSMR NSIMVTAQAR RDVIGPEWGP DGRFTGYASV IGTPDPKPA DIVNKFTVER RPVSNGNFQQ RVKAGDIVVA PYTSDGKITV KLVAGQKDIS STPDYDYRID SSLASSAGFV V AGERWYYT ...String: MNMYKWVPES IRDSGEGQPS YSNNGDYAPS GPWVAAGIHT MPQSLRDSMR NSIMVTAQAR RDVIGPEWGP DGRFTGYASV IGTPDPKPA DIVNKFTVER RPVSNGNFQQ RVKAGDIVVA PYTSDGKITV KLVAGQKDIS STPDYDYRID SSLASSAGFV V AGERWYYT KRHFIIPRYF QNWRMRRRKY VTGWVMPTFY SPKEIFNRLK DSLVPDTGLV TQVWADNNTK RMDFLTAMAE IP QTLSSFL DALGYLGSLI KDFKRRRFFL NKAHQRIRNK LGVSFAERRS QIVSKYDRKI ASARKPAIIV KLRQRKEKAL KAL DKMRVR EEKKMIREFA TQAASLWLSF RYEIMPLYYQ SQDVLDVIAN STSEFMTSRD FVAKAINIGI PLEWNLDQEN LVSQ PRHNV MVKSKLSPEN NIGKTLSVNP FTTAWELLTL SFVVDWFVNF GDVIAGFTGG YSDDSGATAS WRFDDKKVFH LKNIP SAMV IVDINFYTRQ VIDPRLCGGL AFSPKLNLFR YLDAMSLSWN RSRLKISRAT UniProtKB: Maturation protein |
-Macromolecule #3: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 3 / Number of copies: 178 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Acinetobacter phage AP205 (virus) Acinetobacter phage AP205 (virus) |
| Molecular weight | Theoretical: 13.820569 KDa |
| Sequence | String: ANKPMQPITS TANKIVWSDP TRLSTTFSAS LLRQRVKVGI AELNNVSGQY VSVYKRPAPK PEGCADACVI MPNENQSIRT VISGSAENL ATLKAEWETH KRNVDTLFAS GNAGLGFLDP TAAIVSSDTT UniProtKB: Coat protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: 20mM Tris-HCl, 150mM NaCl, pH 8.0 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 100 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III / Details: 3uL sample applied to a 300-mesh 2/1 copper grid. | |||||||||
| Details | AP205 virion particle |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8toc: |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X