-Search query
-Search result
Showing 1 - 50 of 230 items for (author: murphy & ge)

EMDB-42481: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42482: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D

EMDB-42483: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-2heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42485: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42486: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D

PDB-8ur5: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8ur7: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-28570: 
CryoEM structure of PN45545 TCR-CD3 complex
Method: single particle / : Saotome K, Franklin MC
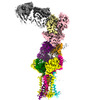
EMDB-28571: 
CryoEM structure of PN45545 TCR-CD3 in complex with HLA-A2 MAGEA4 (230-239)
Method: single particle / : Saotome K, Franklin MC

EMDB-28572: 
CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4
Method: single particle / : Saotome K, Franklin MC
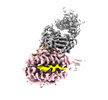
EMDB-28573: 
CryoEM structure of HLA-A2 bound to MAGEA4 (230-239) peptide
Method: single particle / : Saotome K, Franklin MC

EMDB-28574: 
CryoEM structure of HLA-A2 bound to MAGEA8 (232-241) peptide
Method: single particle / : Saotome K, Franklin MC

PDB-8es7: 
CryoEM structure of PN45545 TCR-CD3 complex
Method: single particle / : Saotome K, Franklin MC

PDB-8es8: 
CryoEM structure of PN45545 TCR-CD3 in complex with HLA-A2 MAGEA4 (230-239)
Method: single particle / : Saotome K, Franklin MC

PDB-8es9: 
CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4
Method: single particle / : Saotome K, Franklin MC

PDB-8esa: 
CryoEM structure of HLA-A2 bound to MAGEA4 (230-239) peptide
Method: single particle / : Saotome K, Franklin MC

PDB-8esb: 
CryoEM structure of HLA-A2 bound to MAGEA8 (232-241) peptide
Method: single particle / : Saotome K, Franklin MC

EMDB-27221: 
Cryo-EM map of human LIF signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27227: 
Cryo-EM map of human CNTF signaling complex with full extracellular domains: refined from selected full particle dataset, with better resolution in the interaction core region
Method: single particle / : Zhou Y, Franklin MC

EMDB-27228: 
Cryo-EM structure of human CNTFR alpha in complex with the Fab fragments of two antibodies
Method: single particle / : Zhou Y, Franklin MC

EMDB-27229: 
Cryo-EM map of human CNTF signaling complex with full extracellular domains: refined from a particle subset, with lower global resolution but improved LIFR D6-D8 density
Method: single particle / : Zhou Y, Franklin MC

EMDB-27230: 
Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha
Method: single particle / : Zhou Y, Franklin MC

EMDB-27231: 
Cryo-EM map of human CLCF1 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27244: 
Cryo-EM map of detergent-solubilized human IL-6 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27246: 
Cryo-EM map of human IL-27 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27247: 
Cryo-EM map of human IL-27 signaling complex: focused refinement on the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d6a: 
Cryo-EM structure of human LIF signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d74: 
Cryo-EM structure of human CNTF signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d7e: 
Cryo-EM structure of human CNTFR alpha in complex with the Fab fragments of two antibodies
Method: single particle / : Zhou Y, Franklin MC

PDB-8d7h: 
Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha
Method: single particle / : Zhou Y, Franklin MC

PDB-8d7r: 
Cryo-EM structure of human CLCF1 signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d82: 
Cryo-EM structure of human IL-6 signaling complex in detergent: model containing full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

PDB-8d85: 
Cryo-EM structure of human IL-27 signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC

EMDB-28523: 
Structure of interleukin receptor common gamma chain (IL2Rgamma) in complex with two antibodies
Method: single particle / : Franklin MC, Romero Hernandez A

PDB-8epa: 
Structure of interleukin receptor common gamma chain (IL2Rgamma) in complex with two antibodies
Method: single particle / : Franklin MC, Romero Hernandez A

EMDB-24822: 
20S proteasome from red blood cell lysate
Method: single particle / : Verbeke EJ, Taylor DW

EMDB-26318: 
Influenza Neuraminidase N1-CA09-sNAp-155
Method: single particle / : Acton OJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-26319: 
Influenza Neuraminidase N1-MI15-sNAp-174
Method: single particle / : Acton OJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7u2q: 
Influenza Neuraminidase N1-CA09-sNAp-155
Method: single particle / : Acton OJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7u2t: 
Influenza Neuraminidase N1-MI15-sNAp-174
Method: single particle / : Acton OJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-12638: 
Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit
Method: single particle / : Heit S, Geurts MMG, Murphy BJ, Corey R, Mills DJ, Kuehlbrandt W, Bublitz M

EMDB-12644: 
Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly
Method: single particle / : Heit S, Geurts MMG, Murphy BJ, Corey R, Mills DJ, Kuehlbrandt W, Bublitz M

PDB-7nxf: 
Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit
Method: single particle / : Heit S, Geurts MMG, Murphy BJ, Corey R, Mills DJ, Kuehlbrandt W, Bublitz M

PDB-7ny1: 
Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly
Method: single particle / : Heit S, Geurts MMG, Murphy BJ, Corey R, Mills DJ, Kuehlbrandt W, Bublitz M

EMDB-12196: 
Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase from Methanospirillum hungatei, dimeric complex, conformational state 1, focused refinement of mobile arm and Hdr core
Method: single particle / : Pfeil-Gardiner O, Watanabe T, Shima S, Murphy BJ

EMDB-12197: 
Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei, dimeric complex, focused refinement of conformational state 1 of the mobile arm
Method: single particle / : Pfeil-Gardiner O, Watanabe T, Shima S, Murphy BJ

EMDB-12198: 
Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase from Methanospirillum hungatei, dimeric complex, focused refinement of conformational state 2 of the mobile arm
Method: single particle / : Pfeil-Gardiner O, Watanabe T, Shima S, Murphy BJ

EMDB-12199: 
Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase from Methanospirillum hungatei, dimeric complex, conformational state 2, focused refinement of the mobile arm and Hdr core
Method: single particle / : Pfeil-Gardiner O, Watanabe T, Shima S, Murphy BJ

EMDB-12200: 
Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase from Methanospirillum hungatei, dimeric complex, FdhA subunit by focussed classification and focussed refinement
Method: single particle / : Pfeil-Gardiner O, Watanabe T, Shima S, Murphy BJ

EMDB-12201: 
Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase from Methanospirillum hungatei, dimeric complex, focussed refinement of the FmdABCDG subcomplex
Method: single particle / : Pfeil-Gardiner O, Watanabe T, Shima S, Murphy BJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
