[English] 日本語
 Yorodumi
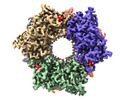
Yorodumi- EMDB-17766: CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Serine protease / Serine protease /  PLANT PROTEIN PLANT PROTEIN | |||||||||
| Function / homology | stem vascular tissue pattern formation / internode patterning / regulation of leaf development / leaf vascular tissue pattern formation / Peptidase S1, PA clan /  nucleoplasm / nucleoplasm /  cytoplasm / Protein NARROW LEAF 1 cytoplasm / Protein NARROW LEAF 1 Function and homology information Function and homology information | |||||||||
| Biological species |   Oryza sativa Japonica Group (Japanese rice) Oryza sativa Japonica Group (Japanese rice) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.4 Å cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Huang LY / Rety S / Xi XG | |||||||||
| Funding support |  China, China,  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2024 Journal: Nat Plants / Year: 2024Title: The catalytic triad of rice NARROW LEAF1 involves H234. Authors: Ling-Yun Huang / Na-Nv Liu / Wei-Fei Chen / Xia Ai / Hai-Hong Li / Ze-Lin Zhang / Xi-Miao Hou / Philippe Fossé / Olivier Mauffret / Dong-Sheng Lei / Stephane Rety / Xu-Guang Xi /   Abstract: NARROW LEAF1 (NAL1) exerts a multifaceted influence on leaf morphology and crop yield. Recent crystal study proposed that histidine 233 (H233) is part of the catalytic triad. Here we report that ...NARROW LEAF1 (NAL1) exerts a multifaceted influence on leaf morphology and crop yield. Recent crystal study proposed that histidine 233 (H233) is part of the catalytic triad. Here we report that unlike suggested previously, H234 instead of H233 is a component of the catalytic triad alongside residues D291 and S385 in NAL1. Remarkably, residue 233 unexpectedly plays a pivotal role in regulating NAL1's proteolytic activity. These findings establish a strong foundation for utilizing NAL1 in breeding programs aimed at improving crop yield. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17766.map.gz emd_17766.map.gz | 24.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17766-v30.xml emd-17766-v30.xml emd-17766.xml emd-17766.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17766_fsc.xml emd_17766_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_17766.png emd_17766.png | 129 KB | ||
| Masks |  emd_17766_msk_1.map emd_17766_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17766.cif.gz emd-17766.cif.gz | 6.4 KB | ||
| Others |  emd_17766_half_map_1.map.gz emd_17766_half_map_1.map.gz emd_17766_half_map_2.map.gz emd_17766_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17766 http://ftp.pdbj.org/pub/emdb/structures/EMD-17766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17766 | HTTPS FTP |
-Related structure data
| Related structure data |  8pn1MC  8pmeC  8pmgC  8pmiC  8pmlC  8pmmC  8pn2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17766.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17766.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.146 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17766_msk_1.map emd_17766_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17766_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17766_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nal1
| Entire | Name: Nal1 |
|---|---|
| Components |
|
-Supramolecule #1: Nal1
| Supramolecule | Name: Nal1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Oryza sativa Japonica Group (Japanese rice) Oryza sativa Japonica Group (Japanese rice) |
-Macromolecule #1: Protein NARROW LEAF 1
| Macromolecule | Name: Protein NARROW LEAF 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Oryza sativa Japonica Group (Japanese rice) Oryza sativa Japonica Group (Japanese rice) |
| Molecular weight | Theoretical: 47.270379 KDa |
| Recombinant expression | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Sequence | String: QSFPCSPSIQ PVASGCTHTE NSAAYFLWPT SNLQHCAAEG RANYFGNLQK GLLPRHPGRL PKGQQANSLL DLMTIRAFHS KILRRFSLG TAVGFRIRKG DLTDIPAILV FVARKVHKKW LNPAQCLPAI LEGPGGVWCD VDVVEFSYYG APAQTPKEQM F SELVDKLC ...String: QSFPCSPSIQ PVASGCTHTE NSAAYFLWPT SNLQHCAAEG RANYFGNLQK GLLPRHPGRL PKGQQANSLL DLMTIRAFHS KILRRFSLG TAVGFRIRKG DLTDIPAILV FVARKVHKKW LNPAQCLPAI LEGPGGVWCD VDVVEFSYYG APAQTPKEQM F SELVDKLC GSDECIGSGS QVASHETFGT LGAIVKRRTG NKQVGFLTNH HVAVDLDYPN QKMFHPLPPN LGPGVYLGAV ER ATSFITD DVWYGIYAGT NPETFVRADG AFIPFADDFD ISTVTTVVRG VGDIGDVKVI DLQCPLNSLI GRQVCKVGRS SGH TTGTVM AYALEYNDEK GICFFTDILV VGENRQTFDL EGDSGSLIIL TSQDGEKPRP IGIIWGGTAN RGRLKLTSDH GPEN WTSGV DLGRLLDRLE LDIIITNESL QDAVQQQR UniProtKB: Protein NARROW LEAF 1 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20mM Tris-HCl 100mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 282 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 50.0 µm / Nominal defocus min: 5.0 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 50.0 µm / Nominal defocus min: 5.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Software | Name: SerialEM (ver. 3.8.4) |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 700 / Average electron dose: 49.02 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)














































