+Search query
-Structure paper
| Title | High-resolution structural-omics of human liver enzymes. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 42, Issue 6, Page 112609, Year 2023 |
| Publish date | Jun 27, 2023 |
 Authors Authors | Chih-Chia Su / Meinan Lyu / Zhemin Zhang / Masaru Miyagi / Wei Huang / Derek J Taylor / Edward W Yu /  |
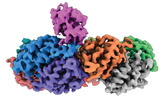
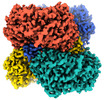
| PubMed Abstract | We applied raw human liver microsome lysate to a holey carbon grid and used cryo-electron microscopy (cryo-EM) to define its composition. From this sample we identified and simultaneously determined ...We applied raw human liver microsome lysate to a holey carbon grid and used cryo-electron microscopy (cryo-EM) to define its composition. From this sample we identified and simultaneously determined high-resolution structural information for ten unique human liver enzymes involved in diverse cellular processes. Notably, we determined the structure of the endoplasmic bifunctional protein H6PD, where the N- and C-terminal domains independently possess glucose-6-phosphate dehydrogenase and 6-phosphogluconolactonase enzymatic activity, respectively. We also obtained the structure of heterodimeric human GANAB, an ER glycoprotein quality-control machinery that contains a catalytic α subunit and a noncatalytic β subunit. In addition, we observed a decameric peroxidase, PRDX4, which directly contacts a disulfide isomerase-related protein, ERp46. Structural data suggest that several glycosylations, bound endogenous compounds, and ions associate with these human liver enzymes. These results highlight the importance of cryo-EM in facilitating the elucidation of human organ proteomics at the atomic level. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:37289586 / PubMed:37289586 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.64 - 3.47 Å |
| Structure data | EMDB-26915, PDB-7uzm: EMDB-28214, PDB-8ekw: EMDB-28217, PDB-8eky: EMDB-28232, PDB-8em2: EMDB-28262, PDB-8emr: EMDB-28263, PDB-8ems: EMDB-28264, PDB-8emt: EMDB-28271, PDB-8ene: EMDB-28377, PDB-8eoj: EMDB-28465, PDB-8eor: |
| Chemicals |  ChemComp-HOH:  ChemComp-NAG:  ChemComp-CA:  ChemComp-PLP:  ChemComp-FES:  ChemComp-MTE:  ChemComp-MOS:  ChemComp-FAD: 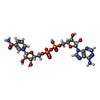 ChemComp-NAD:  ChemComp-EEE: |
| Source |
|
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  aldehyde dehydrogenase / aldehyde dehydrogenase /  human liver / human liver /  Peroxiredoxin-4 / H6PD / Peroxiredoxin-4 / H6PD /  HYDROLASE / HYDROLASE /  glucosidase II / GANAB / glucosidase II / GANAB /  glycosyl hydrolase 31 family / glycosyl hydrolase 31 family /  Glycogen Phosphorylase / PLP / Glycogen Phosphorylase / PLP /  Aldehyde oxidase / AOX1 / Aldehyde oxidase / AOX1 /  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  Microsomal triglyceride transfer protein / LIPID TRANSPORT / Microsomal triglyceride transfer protein / LIPID TRANSPORT /  ISOMERASE / ISOMERASE /  carboxylesterase carboxylesterase |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers