+Search query
-Structure paper
| Title | Transport and inhibition mechanisms of human VMAT2. |
|---|---|
| Journal, issue, pages | Nature, Vol. 626, Issue 7998, Page 427-434, Year 2024 |
| Publish date | Dec 11, 2023 |
 Authors Authors | Di Wu / Qihao Chen / Zhuoya Yu / Bo Huang / Jun Zhao / Yuhang Wang / Jiawei Su / Feng Zhou / Rui Yan / Na Li / Yan Zhao / Daohua Jiang /  |
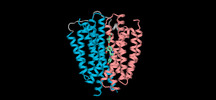
| PubMed Abstract | Vesicular monoamine transporter 2 (VMAT2) accumulates monoamines in presynaptic vesicles for storage and exocytotic release, and has a vital role in monoaminergic neurotransmission. Dysfunction of ...Vesicular monoamine transporter 2 (VMAT2) accumulates monoamines in presynaptic vesicles for storage and exocytotic release, and has a vital role in monoaminergic neurotransmission. Dysfunction of monoaminergic systems causes many neurological and psychiatric disorders, including Parkinson's disease, hyperkinetic movement disorders and depression. Suppressing VMAT2 with reserpine and tetrabenazine alleviates symptoms of hypertension and Huntington's disease, respectively. Here we describe cryo-electron microscopy structures of human VMAT2 complexed with serotonin and three clinical drugs at 3.5-2.8 Å, demonstrating the structural basis for transport and inhibition. Reserpine and ketanserin occupy the substrate-binding pocket and lock VMAT2 in cytoplasm-facing and lumen-facing states, respectively, whereas tetrabenazine binds in a VMAT2-specific pocket and traps VMAT2 in an occluded state. The structures in three distinct states also reveal the structural basis of the VMAT2 transport cycle. Our study establishes a structural foundation for the mechanistic understanding of substrate recognition, transport, drug inhibition and pharmacology of VMAT2 while shedding light on the rational design of potential therapeutic agents. |
 External links External links |  Nature / Nature /  PubMed:38081299 PubMed:38081299 |
| Methods | EM (single particle) |
| Resolution | 2.84 - 3.52 Å |
| Structure data | EMDB-36628, PDB-8jsw: EMDB-36637, PDB-8jt9: EMDB-36638, PDB-8jta: EMDB-36640, PDB-8jtc: |
| Chemicals |  ChemComp-SRO:  ChemComp-UYX:  ChemComp-YHL:  ChemComp-YHR: |
| Source |
|
 Keywords Keywords |  TRANSPORT PROTEIN / transporterA / transporter1 / transporter2 TRANSPORT PROTEIN / transporterA / transporter1 / transporter2 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers