+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human VMAT2 complex with ketanserin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter1 /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationserotonin secretion by mast cell / sequestering of neurotransmitter / somato-dendritic dopamine secretion / histamine uptake / neurotransmitter loading into synaptic vesicle / monoamine:proton antiporter activity / aminergic neurotransmitter loading into synaptic vesicle / clathrin-sculpted monoamine transport vesicle membrane / Serotonin Neurotransmitter Release Cycle / serotonin:sodium:chloride symporter activity ...serotonin secretion by mast cell / sequestering of neurotransmitter / somato-dendritic dopamine secretion / histamine uptake / neurotransmitter loading into synaptic vesicle / monoamine:proton antiporter activity / aminergic neurotransmitter loading into synaptic vesicle / clathrin-sculpted monoamine transport vesicle membrane / Serotonin Neurotransmitter Release Cycle / serotonin:sodium:chloride symporter activity / Dopamine Neurotransmitter Release Cycle / Norepinephrine Neurotransmitter Release Cycle / serotonin uptake / dopaminergic synapse / Na+/Cl- dependent neurotransmitter transporters / monoamine transmembrane transporter activity / dopamine transport / monoamine transport / histamine secretion by mast cell /  neurotransmitter transport / negative regulation of reactive oxygen species biosynthetic process / response to amphetamine / post-embryonic development / secretory granule membrane / locomotory behavior / neurotransmitter transport / negative regulation of reactive oxygen species biosynthetic process / response to amphetamine / post-embryonic development / secretory granule membrane / locomotory behavior /  terminal bouton / synaptic vesicle membrane / response to toxic substance / terminal bouton / synaptic vesicle membrane / response to toxic substance /  synaptic vesicle / chemical synaptic transmission / synaptic vesicle / chemical synaptic transmission /  axon / intracellular membrane-bounded organelle / axon / intracellular membrane-bounded organelle /  centrosome / centrosome /  dendrite / dendrite /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.97 Å cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | Jiang DH / Wu D | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Transport and inhibition mechanisms of human VMAT2. Authors: Di Wu / Qihao Chen / Zhuoya Yu / Bo Huang / Jun Zhao / Yuhang Wang / Jiawei Su / Feng Zhou / Rui Yan / Na Li / Yan Zhao / Daohua Jiang /  Abstract: Vesicular monoamine transporter 2 (VMAT2) accumulates monoamines in presynaptic vesicles for storage and exocytotic release, and has a vital role in monoaminergic neurotransmission. Dysfunction of ...Vesicular monoamine transporter 2 (VMAT2) accumulates monoamines in presynaptic vesicles for storage and exocytotic release, and has a vital role in monoaminergic neurotransmission. Dysfunction of monoaminergic systems causes many neurological and psychiatric disorders, including Parkinson's disease, hyperkinetic movement disorders and depression. Suppressing VMAT2 with reserpine and tetrabenazine alleviates symptoms of hypertension and Huntington's disease, respectively. Here we describe cryo-electron microscopy structures of human VMAT2 complexed with serotonin and three clinical drugs at 3.5-2.8 Å, demonstrating the structural basis for transport and inhibition. Reserpine and ketanserin occupy the substrate-binding pocket and lock VMAT2 in cytoplasm-facing and lumen-facing states, respectively, whereas tetrabenazine binds in a VMAT2-specific pocket and traps VMAT2 in an occluded state. The structures in three distinct states also reveal the structural basis of the VMAT2 transport cycle. Our study establishes a structural foundation for the mechanistic understanding of substrate recognition, transport, drug inhibition and pharmacology of VMAT2 while shedding light on the rational design of potential therapeutic agents. | |||||||||
| History |
|
- Structure visualization
Structure visualization
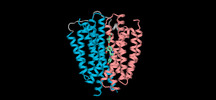
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36637.map.gz emd_36637.map.gz | 116 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36637-v30.xml emd-36637-v30.xml emd-36637.xml emd-36637.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36637.png emd_36637.png | 44.3 KB | ||
| Filedesc metadata |  emd-36637.cif.gz emd-36637.cif.gz | 5.4 KB | ||
| Others |  emd_36637_half_map_1.map.gz emd_36637_half_map_1.map.gz emd_36637_half_map_2.map.gz emd_36637_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36637 http://ftp.pdbj.org/pub/emdb/structures/EMD-36637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36637 | HTTPS FTP |
-Related structure data
| Related structure data |  8jt9MC  8jswC  8jtaC  8jtcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36637.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36637.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36637_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
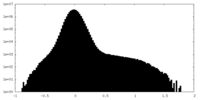
| Density Histograms |
-Half map: #1
| File | emd_36637_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : transporter1
| Entire | Name: transporter1 |
|---|---|
| Components |
|
-Supramolecule #1: transporter1
| Supramolecule | Name: transporter1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Synaptic vesicular amine transporter
| Macromolecule | Name: Synaptic vesicular amine transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.086848 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SRKLILFIVF LALLLDNMLL TVVVPIIPSY LYSIKHEKNA TEIQTARPVH TASISDSFQS IFSYYDNSTM VTGNATRDLT LHQTATQHM VTNASAVPSD CPSEDKDLLN ENVQVGLLFA SKATVQLITN PFIGLLTNRI GYPIPIFAGF CIMFVSTIMF A FSSSYAFL ...String: SRKLILFIVF LALLLDNMLL TVVVPIIPSY LYSIKHEKNA TEIQTARPVH TASISDSFQS IFSYYDNSTM VTGNATRDLT LHQTATQHM VTNASAVPSD CPSEDKDLLN ENVQVGLLFA SKATVQLITN PFIGLLTNRI GYPIPIFAGF CIMFVSTIMF A FSSSYAFL LIARSLQGIG SSCSSVAGMG MLASVYTDDE ERGNVMGIAL GGLAMGVLVG PPFGSVLYEF VGKTAPFLVL AA LVLLDGA IQLFVLQPSR VQPESQKGTP LTTLLKDPYI LIAAGSICFA NMGIAMLEPA LPIWMMETMC SRKWQLGVAF LPA SISYLI GTNIFGILAH KMGRWLCALL GMIIVGVSIL CIPFAKNIYG LIAPNFGVGF AIGMVDSSMM PIMGYLVDLR HVSV YGSVY AIADVAFCMG YAIGPSAGGA IAKAIGFPWL MTIIGIIDIL FAPLCFFLRS PP UniProtKB: Synaptic vesicular amine transporter |
-Macromolecule #2: 3-[2-[4-(4-fluorophenyl)carbonylpiperidin-1-yl]ethyl]-1~{H}-quina...
| Macromolecule | Name: 3-[2-[4-(4-fluorophenyl)carbonylpiperidin-1-yl]ethyl]-1~{H}-quinazoline-2,4-dione type: ligand / ID: 2 / Number of copies: 1 / Formula: UYX |
|---|---|
| Molecular weight | Theoretical: 395.427 Da |
| Chemical component information |  ChemComp-UYX: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.97 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 504617 |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X