-Search query
-Search result
Showing 1 - 50 of 124 items for (author: lei & ht)

EMDB-40180: 
MsbA bound to cerastecin C
Method: single particle / : Chen Y, Klein D

EMDB-18482: 
Herpes simplex virus 1 capsid (WT) vertices in perinuclear NEC-coated vesicles determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18484: 
Herpes simplex virus 1 nuclear egress complex (WT) determined in situ from perinuclear vesicles
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-17974: 
Pseudorabies virus cytosolic C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-17975: 
Pseudorabies virus primary enveloped (perinuclear) C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-17976: 
Pseudorabies nuclear C-capsids (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18473: 
Subtomogram average of pseudorabies virus nuclear egress complex helical form (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18474: 
Subtomogram average of pseudorabies virus nuclear egress complex (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18479: 
Pseudorabies virus cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18480: 
Pseudorabies virus nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18481: 
Herpes simplex virus 1 cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D
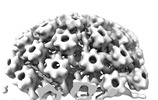
EMDB-18483: 
Herpes simplex virus 1 nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-27641: 
The structure of the interleukin 11 signalling complex, truncated gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW

EMDB-27642: 
The structure of the IL-11 signalling complex, with full-length extracellular gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW

PDB-8dps: 
The structure of the interleukin 11 signalling complex, truncated gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW

PDB-8dpt: 
The structure of the IL-11 signalling complex, with full-length extracellular gp130
Method: single particle / : Metcalfe RD, Hanssen E, Griffin MDW

EMDB-16458: 
Electron cryo-tomography and subtomogram averaging of cytoplasmic lattice filaments from mammalian oocytes
Method: subtomogram averaging / : Petrovic A, Bauerlein FJB, Jentoft IMA, Schuh M, Fernandez-Busnadiego R

EMDB-16472: 
In situ cryo-electron tomogram of cytoplasmic lattice filaments from a mouse oocyte
Method: electron tomography / : Bauerlein FJB, Jentoft IMA, Petrovic A, Fernandez-Busnadiego R, Schuh M

EMDB-41158: 
CS2it1p2_F7K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-41180: 
Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab
Method: single particle / : Goldsmith JG, McLellan JS

EMDB-34813: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril
Method: helical / : Low JYK, Pervushin K

PDB-8hia: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril
Method: helical / : Low JYK, Pervushin K

EMDB-33845: 
Cryo-EM map of Rpd3S complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33846: 
Cryo-EM map of Rpd3S:head-bridge-right arm
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33847: 
Cryo-EM map of Rpd3S:bridge-left arm
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33848: 
Cryo-EM map of Eaf3 CHD bound to H3K36me3 nucleosome
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33849: 
Cryo-EM map of Rpd3S in loose-state Rpd3S-NCP complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33850: 
Cryo-EM map of Rpd3S in close-state Rpd3S-NCP complex
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33851: 
Cryo-EM map of Rpd3S complex bound to H3K36me3 nucleosome in close state
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-33852: 
Cryo-EM msp of Rpd3S complex bound to H3K36me3 nucleosome in loose state
Method: single particle / : Li HT, Yan CY, Guan HP, Wang P

EMDB-27148: 
Zebrafish MFSD2A isoform B in inward open ligand bound conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27149: 
Zebrafish MFSD2A isoform B in inward open ligand-free conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27150: 
Zebrafish MFSD2A isoform B in inward open ligand 1A conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27151: 
Zebrafish MFSD2A isoform B in inward open ligand 1B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27152: 
Zebrafish MFSD2A isoform B in inward open ligand 2B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27153: 
Zebrafish MFSD2A isoform B in inward open ligand 3C conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-16128: 
Tomogram of a late endosome of A549 cell infected with influenza A virus.
Method: electron tomography / : Klein S, Chlanda P

EMDB-16129: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6C).
Method: electron tomography / : Klein S, Chlanda P

EMDB-16130: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6E)
Method: electron tomography / : Klein S, Chlanda P

EMDB-16131: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6G)
Method: electron tomography / : Klein S, Chlanda P

EMDB-16132: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6I,K,M)
Method: electron tomography / : Klein S, Chlanda P

EMDB-16133: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6O)
Method: electron tomography / : Klein S, Chlanda P

EMDB-28092: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28090: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-040
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO

EMDB-28091: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-045
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO
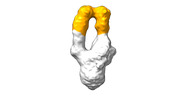
EMDB-28093: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156
Method: single particle / : Shek J, Callaway H, Li H, Yu X, Saphire EO

EMDB-27243: 
Cryo-EM map of MORF-WHs in complex with 197bp nucleosome aided by scFv
Method: single particle / : Zhou BR
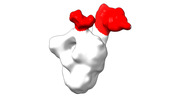
EMDB-28094: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-234
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28095: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-260
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
