+Search query
-Structure paper
| Title | Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii. |
|---|---|
| Journal, issue, pages | Nat Microbiol, Vol. 9, Issue 5, Page 1244-1255, Year 2024 |
| Publish date | Apr 22, 2024 |
 Authors Authors | Hao Wang / Andrii Ishchenko / Jason Skudlarek / Pamela Shen / Liudmila Dzhekieva / Ronald E Painter / Yun-Ting Chen / Marina Bukhtiyarova / Andrew Leithead / Rodger Tracy / Kerim Babaoglu / Carolyn Bahnck-Teets / Alexei Buevich / Tamara D Cabalu / Marc Labroli / Henry Lange / Ying Lei / Wei Li / Jian Liu / Paul A Mann / Tao Meng / Helen J Mitchell / James Mulhearn / Giovanna Scapin / Deyou Sha / Anthony W Shaw / Qian Si / Ling Tong / Chengwei Wu / Zhe Wu / Jing Chen Xiao / Min Xu / Li-Kang Zhang / David McKenney / Randy R Miller / Todd A Black / Andrew Cooke / Carl J Balibar / Daniel J Klein / Izzat Raheem / Scott S Walker /  |
| PubMed Abstract | Carbapenem-resistant Acinetobacter baumannii infections have limited treatment options. Synthesis, transport and placement of lipopolysaccharide or lipooligosaccharide (LOS) in the outer membrane of ...Carbapenem-resistant Acinetobacter baumannii infections have limited treatment options. Synthesis, transport and placement of lipopolysaccharide or lipooligosaccharide (LOS) in the outer membrane of Gram-negative bacteria are important for bacterial virulence and survival. Here we describe the cerastecins, inhibitors of the A. baumannii transporter MsbA, an LOS flippase. These molecules are potent and bactericidal against A. baumannii, including clinical carbapenem-resistant Acinetobacter baumannii isolates. Using cryo-electron microscopy and biochemical analysis, we show that the cerastecins adopt a serpentine configuration in the central vault of the MsbA dimer, stalling the enzyme and uncoupling ATP hydrolysis from substrate flipping. A derivative with optimized potency and pharmacokinetic properties showed efficacy in murine models of bloodstream or pulmonary A. baumannii infection. While resistance development is inevitable, targeting a clinically unexploited mechanism avoids existing antibiotic resistance mechanisms. Although clinical validation of LOS transport remains undetermined, the cerastecins may open a path to narrow-spectrum treatment modalities for important nosocomial infections. |
 External links External links |  Nat Microbiol / Nat Microbiol /  PubMed:38649414 PubMed:38649414 |
| Methods | EM (single particle) |
| Resolution | 3.32 Å |
| Structure data | EMDB-40180, PDB-8gk7: |
| Chemicals | 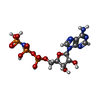 ChemComp-ANP: 
ChemComp-ZQF: |
| Source |
|
 Keywords Keywords |  ANTIMICROBIAL PROTEIN / ANTIMICROBIAL PROTEIN /  MsbA / MsbA /  antibacterial / antibacterial /  inhibitor / cerastecin inhibitor / cerastecin |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






