-Search query
-Search result
Showing 1 - 50 of 3,277 items for (author: thom & c)

EMDB-17449: 
S. cerevisiae nexus-sCMGE after DNA replication initiation
Method: single particle / : Henrikus SS, Willhoft O

EMDB-17458: 
S. cerevisiae ssDNA-sCMGE after DNA replication initiation
Method: single particle / : Henrikus SS, Willhoft O

EMDB-17459: 
S. cerevisiae consensus-sCMGE on ssDNA after DNA replication initiation
Method: single particle / : Henrikus SS, Willhoft O

EMDB-17460: 
S. cerevisiae sCMGE with N-ter Mcm10 density
Method: single particle / : Henrikus SS, Willhoft O

PDB-8p5e: 
S. cerevisiae nexus-sCMGE after DNA replication initiation
Method: single particle / : Henrikus SS, Willhoft O

PDB-8p62: 
S. cerevisiae ssDNA-sCMGE after DNA replication initiation
Method: single particle / : Henrikus SS, Willhoft O

PDB-8p63: 
S. cerevisiae consensus-sCMGE on ssDNA after DNA replication initiation
Method: single particle / : Henrikus SS, Willhoft O

EMDB-19066: 
TREK2 in OGNG/CHS detergent micelle with biparatopic inhibitory nanobody Nb6158
Method: single particle / : Smith KHM, Tucker SJ

EMDB-19854: 
PHF type tau filament from R406W mutant
Method: helical / : Qi C, Scheres SHW, Michel G

EMDB-19855: 
PHF type tau filament from in vitro V337M mutant
Method: helical / : Qi C, Lovestam S, Scheres SHW, Michel G

PDB-9eog: 
PHF type tau filament from R406W mutant
Method: helical / : Qi C, Scheres SHW, Michel G

PDB-9eoh: 
PHF type tau filament from in vitro V337M mutant
Method: helical / : Qi C, Lovestam S, Scheres SHW, Michel G

EMDB-41271: 
Consensus map of 96nm repeat of human respiratory doublet microtubule, RS1-2 region
Method: single particle / : Gui M, Brown A

EMDB-19798: 
human PLD3 homodimer structure
Method: single particle / : Lammens K

EMDB-18639: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)
Method: single particle / : Ren J, Stuart DI, Duyvesteyn HME

EMDB-18649: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
Method: single particle / : Ren J, Duyvesteyn HME, Stuart DI

EMDB-19002: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8qsq: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)
Method: single particle / : Ren J, Stuart DI, Duyvesteyn HME

PDB-8qtd: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
Method: single particle / : Ren J, Duyvesteyn HME, Stuart DI

PDB-8r8k: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-43683: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F

EMDB-43684: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F

PDB-8vzn: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F

PDB-8vzo: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F
EMDB-41373: 
E. coli MraY mutant-T23P
Method: single particle / : Orta AK, Li YE, Clemons WM

PDB-8tlu: 
E. coli MraY mutant-T23P
Method: single particle / : Orta AK, Li YE, Clemons WM

EMDB-43751: 
TRPM7 structure in complex with anticancer agent CCT128930 in closed state
Method: single particle / : Nadezhdin KD, Sobolevsky AI

PDB-8w2l: 
TRPM7 structure in complex with anticancer agent CCT128930 in closed state
Method: single particle / : Nadezhdin KD, Sobolevsky AI

EMDB-41151: 
CryoEM structure of nucleotide-free form of the nitrogenase iron protein from A. vinelandii
Method: single particle / : Warmack RA, Rees DC

PDB-8tc3: 
CryoEM structure of nucleotide-free form of the nitrogenase iron protein from A. vinelandii
Method: single particle / : Warmack RA, Rees DC

EMDB-19735: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, basal state, helical reconstruction
Method: helical / : McCorvie TJ, Yue WW

EMDB-19736: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, basal state, single particle reconstruction
Method: single particle / : McCorvie TJ, Yue WW

EMDB-19737: 
Full-length human cystathionine beta-synthase, basal state, helical reconstruction
Method: helical / : McCorvie TJ, Yue WW

EMDB-19738: 
Full-length human cystathionine beta-synthase, basal state, single particle reconstruction
Method: single particle / : McCorvie TJ, Yue WW

EMDB-19739: 
Full-length human cystathionine beta-synthase, basal state, partially degraded tetramer
Method: single particle / : McCorvie TJ, Yue WW
EMDB-19740: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, SAM bound, activated state, helical reconstruction
Method: helical / : McCorvie TJ, Yue WW

EMDB-19741: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, SAM bound, activated state, local helical reconstruction
Method: helical / : McCorvie TJ, Yue WW

EMDB-19742: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, SAM bound, activated state, local single particle reconstruction
Method: single particle / : McCorvie TJ, Yue WW

PDB-8s5h: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, basal state, helical reconstruction
Method: helical / : McCorvie TJ, Yue WW

PDB-8s5i: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, basal state, single particle reconstruction
Method: single particle / : McCorvie TJ, Yue WW

PDB-8s5j: 
Full-length human cystathionine beta-synthase, basal state, helical reconstruction
Method: helical / : McCorvie TJ, Yue WW

PDB-8s5k: 
Full-length human cystathionine beta-synthase, basal state, single particle reconstruction
Method: single particle / : McCorvie TJ, Yue WW

PDB-8s5l: 
Full-length human cystathionine beta-synthase, basal state, partially degraded tetramer
Method: single particle / : McCorvie TJ, Yue WW

PDB-8s5m: 
Full-length human cystathionine beta-synthase with C-terminal 6xHis-tag, SAM bound, activated state, helical reconstruction
Method: helical / : McCorvie TJ, Yue WW

EMDB-17863: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod
Method: helical / : Hospenthal M, Zyla D, Glockshuber R, Waksman G

EMDB-17878: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod
Method: helical / : Zyla D, Hospenthal M, Glockshuber R, Waksman G

PDB-8psv: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod
Method: helical / : Hospenthal M, Zyla D, Glockshuber R, Waksman G

PDB-8ptu: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod
Method: helical / : Zyla D, Hospenthal M, Glockshuber R, Waksman G
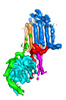
EMDB-40572: 
Human adenylyl Cyclase 5 in complex with Gbg
Method: single particle / : Yen YC, Tesmer JJG
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
