+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli MraY mutant-T23P | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  peptidoglycan / peptidoglycan /  phosphotransferase / phosphotransferase /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information phospho-N-acetylmuramoyl-pentapeptide-transferase / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity / phospho-N-acetylmuramoyl-pentapeptide-transferase / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity /  phospho-N-acetylmuramoyl-pentapeptide-transferase activity / cell wall macromolecule biosynthetic process / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape / phospho-N-acetylmuramoyl-pentapeptide-transferase activity / cell wall macromolecule biosynthetic process / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape /  cell cycle / cell cycle /  cell division / cell division /  membrane ... membrane ... phospho-N-acetylmuramoyl-pentapeptide-transferase / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity / phospho-N-acetylmuramoyl-pentapeptide-transferase / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity /  phospho-N-acetylmuramoyl-pentapeptide-transferase activity / cell wall macromolecule biosynthetic process / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape / phospho-N-acetylmuramoyl-pentapeptide-transferase activity / cell wall macromolecule biosynthetic process / peptidoglycan biosynthetic process / cell wall organization / regulation of cell shape /  cell cycle / cell cycle /  cell division / cell division /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) | |||||||||
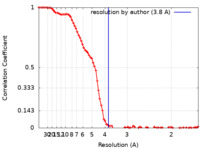
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Orta AK / Li YE / Clemons WM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa. Authors: Lindsey S Marmont / Anna K Orta / Becca W A Baileeves / David Sychantha / Ana Fernández-Galliano / Yancheng E Li / Neil G Greene / Robin A Corey / Phillip J Stansfeld / William M Clemons / ...Authors: Lindsey S Marmont / Anna K Orta / Becca W A Baileeves / David Sychantha / Ana Fernández-Galliano / Yancheng E Li / Neil G Greene / Robin A Corey / Phillip J Stansfeld / William M Clemons / Thomas G Bernhardt /    Abstract: Many bacterial surface glycans such as the peptidoglycan (PG) cell wall are built from monomeric units linked to a polyprenyl lipid carrier. How this limiting carrier is distributed among competing ...Many bacterial surface glycans such as the peptidoglycan (PG) cell wall are built from monomeric units linked to a polyprenyl lipid carrier. How this limiting carrier is distributed among competing pathways has remained unclear. Here we describe the isolation of hyperactive variants of Pseudomonas aeruginosa MraY, the enzyme that forms the first lipid-linked PG precursor. These variants result in the elevated production of the final PG precursor lipid II in cells and are hyperactive in vitro. The activated MraY variants have substitutions that map to a cavity on the extracellular side of the dimer interface, far from the active site. Our structural and molecular dynamics results suggest that this cavity is a binding site for externalized lipid II. Overall, our results support a model in which excess externalized lipid II allosterically inhibits MraY, providing a feedback mechanism that prevents the sequestration of lipid carrier in the PG biogenesis pathway. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41373.map.gz emd_41373.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41373-v30.xml emd-41373-v30.xml emd-41373.xml emd-41373.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41373_fsc.xml emd_41373_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_41373.png emd_41373.png | 103.2 KB | ||
| Masks |  emd_41373_msk_1.map emd_41373_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41373.cif.gz emd-41373.cif.gz | 6.1 KB | ||
| Others |  emd_41373_additional_1.map.gz emd_41373_additional_1.map.gz emd_41373_half_map_1.map.gz emd_41373_half_map_1.map.gz emd_41373_half_map_2.map.gz emd_41373_half_map_2.map.gz | 49.6 MB 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41373 http://ftp.pdbj.org/pub/emdb/structures/EMD-41373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41373 | HTTPS FTP |
-Related structure data
| Related structure data |  8tluMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41373.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41373.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41373_msk_1.map emd_41373_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map
| File | emd_41373_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41373_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41373_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli MraY T23P dimer
| Entire | Name: E. coli MraY T23P dimer |
|---|---|
| Components |
|
-Supramolecule #1: E. coli MraY T23P dimer
| Supramolecule | Name: E. coli MraY T23P dimer / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 39.875 KDa |
-Macromolecule #1: Phospho-N-acetylmuramoyl-pentapeptide-transferase
| Macromolecule | Name: Phospho-N-acetylmuramoyl-pentapeptide-transferase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 39.905551 KDa |
| Recombinant expression | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Sequence | String: MLVWLAEHLV KYYSGFNVFS YLPFRAIVSL LTALFISLWM GPRMIAHLQK LSFGQVVRND GPESHFSKRG TPTMGGIMIL TAIVISVLL WAYPSNPYVW CVLVVLVGYG VIGFVDDYRK VVRKDTKGLI ARWKYFWMSV IALGVAFALY LAGKDTPATQ L VVPFFKDV ...String: MLVWLAEHLV KYYSGFNVFS YLPFRAIVSL LTALFISLWM GPRMIAHLQK LSFGQVVRND GPESHFSKRG TPTMGGIMIL TAIVISVLL WAYPSNPYVW CVLVVLVGYG VIGFVDDYRK VVRKDTKGLI ARWKYFWMSV IALGVAFALY LAGKDTPATQ L VVPFFKDV MPQLGLFYIL LAYFVIVGTG NAVNLTDGLD GLAIMPTVFV AGGFALVAWA TGNMNFASYL HIPYLRHAGE LV IVCTAIV GAGLGFLWFN TYPAQVFMGD VGSLALGGAL GIIAVLLRQE FLLVIMGGVF VVETLSVILQ VGSFKLRGQR IFR MAPIHH HYELKGWPEP RVIVRFWIIS LMLVLIGLAT LKVR UniProtKB:  Phospho-N-acetylmuramoyl-pentapeptide-transferase Phospho-N-acetylmuramoyl-pentapeptide-transferase |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | MraY was pulled down using the YES complex. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 Bright-field microscopy / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X