-Search query
-Search result
Showing 1 - 50 of 95 items for (author: schwartz & e)

EMDB-18658: 
Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex
Method: single particle / : Hoelzgen F, Klukin E, Zalk R, Shahar A, Cohen-Schwartz S, Frank GA

PDB-8qu9: 
Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex
Method: single particle / : Hoelzgen F, Klukin E, Zalk R, Shahar A, Cohen-Schwartz S, Frank GA

EMDB-40248: 
CRISPR-Cas type III-D effector complex
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40250: 
CRISPR-Cas type III-D effector complex bound to a self-target RNA in the pre-cleavage state
Method: single particle / : Schwartz EA, Taylor DW
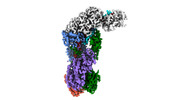
EMDB-40251: 
CRISPR-Cas type III-D effector complex bound to self-target RNA in a post-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40276: 
CRISPR-Cas type III-D effector complex consensus map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40296: 
CRISPR-Cas type III-D effector complex local refinement map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40297: 
CRISPR-Cas type III-D effector complex bound to a target RNA local refinement map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40298: 
CRISPR-Cas type III-D effector complex bound to a target RNA consensus map
Method: single particle / : Schwartz EA, Taylor DW

PDB-8s9t: 
CRISPR-Cas type III-D effector complex
Method: single particle / : Schwartz EA, Taylor DW

PDB-8s9v: 
CRISPR-Cas type III-D effector complex bound to a self-target RNA in the pre-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

PDB-8s9x: 
CRISPR-Cas type III-D effector complex bound to self-target RNA in a post-cleavage state
Method: single particle / : Schwartz EA, Taylor DW
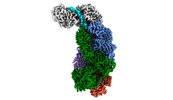
EMDB-40249: 
CRISPR-Cas type III-D effector complex bound to a target RNA
Method: single particle / : Schwartz EA, Taylor DW

PDB-8s9u: 
CRISPR-Cas type III-D effector complex bound to a target RNA
Method: single particle / : Schwartz EA, Taylor DW

EMDB-17208: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17212: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-17249: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17250: 
CRYO-EM FOCUSED REFINEMENT MAPS OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17254: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 SKO
Method: single particle / : RAJAN KS, YONATH A

EMDB-17255: 
CRYO-EM FOCUSED REFINEMENT OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

PDB-8ova: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8ove: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-14248: 
Chlamydomonas reinhardtii TSP9 mutant small Photosystem I complex
Method: single particle / : Klaiman D, Schwartz T, Nelson N

PDB-7r3k: 
Chlamydomonas reinhardtii TSP9 mutant small Photosystem I complex
Method: single particle / : Klaiman D, Schwartz T, Nelson N

EMDB-28089: 
a22L prion fibril
Method: helical / : Hoyt F, Caughey B

EMDB-14734: 
Tomogram of human sperm singlet microtubules
Method: electron tomography / : Hoog JL, Zabeo DZ

EMDB-15450: 
TAILS structure in human sperm tip singlet microtubules
Method: subtomogram averaging / : Hoog JL, Zabeo DZ

EMDB-25824: 
aRML prion fibril
Method: single particle / : Hoyt F, Standke HG, Artikis E, Caughey B, Kraus A

PDB-7td6: 
aRML prion fibril
Method: single particle / : Hoyt F, Standke HG, Artikis E, Caughey B, Kraus A
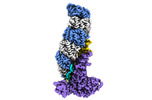
EMDB-24974: 
Structure of type I-D Cascade bound to a dsDNA target
Method: single particle / : Schwartz EA, Taylor DW
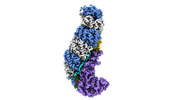
EMDB-24976: 
Structure of type I-D Cascade bound to a ssRNA target
Method: single particle / : Schwartz EA, Taylor DW

PDB-7sba: 
Structure of type I-D Cascade bound to a dsDNA target
Method: single particle / : Schwartz EA, Taylor DW

PDB-7sbb: 
Structure of type I-D Cascade bound to a ssRNA target
Method: single particle / : Schwartz EA, Taylor DW

EMDB-14853: 
SARS-CoV-2 Spike in complex with the neutralizing antibody Cv2.1169
Method: single particle / : Guardado-Calvo P, Fernandez I, Rey FA

EMDB-13967: 
Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 4
Method: single particle / : Rebelo-Guiomar P, Pellegrino S, Dent KC, Warren AJ, Minczuk M

PDB-7qh7: 
Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 4
Method: single particle / : Rebelo-Guiomar P, Pellegrino S, Dent KC, Warren AJ, Minczuk M

EMDB-24876: 
SARS-CoV-2-6P-Mut2 S protein
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-24877: 
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 1)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-24878: 
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 2)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-24879: 
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (conformation 3)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-26389: 
J08 fragment antigen binding in complex with Omicron SARS-CoV-2-6P S protein
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-7s6i: 
SARS-CoV-2-6P-Mut2 S protein
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-7s6j: 
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 1)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-7s6k: 
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 2)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-7s6l: 
J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (conformation 3)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-13962: 
Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 2
Method: single particle / : Rebelo-Guiomar P, Pellegrino S, Dent KC, Warren AJ, Minczuk M

EMDB-13963: 
Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 3
Method: single particle / : Rebelo-Guiomar P, Pellegrino S, Dent KC, Warren AJ, Minczuk M

EMDB-13965: 
Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 1
Method: single particle / : Rebelo-Guiomar P, Pellegrino S, Dent KC, Warren AJ, Minczuk M

EMDB-13966: 
Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 5
Method: single particle / : Rebelo-Guiomar P, Pellegrino S, Dent KC, Warren AJ, Minczuk M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
