-Search query
-Search result
Showing 1 - 50 of 84 items for (author: liu & yy)

EMDB-35878: 
Cryo-EM structure of DIP-2I8I polymorph 2
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35816: 
Cryo-EM structure of DIP-2I8I fibril polymorph1
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-40799: 
Cryo-EM consensus map of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

EMDB-35519: 
Cryo-EM structure of hnRAC1-8I fibril
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35520: 
Cryo-EM structure of hnRAC1-2I8I fibril.
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35508: 
Cryo-EM structure of hnRAC1-2I fibril.
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35706: 
Cryo-EM structure of GIPR splice variant 1 (SV1) in complex with Gs protein
Method: single particle / : Zhao FH, Hang KN, Zhou QT, Shao LJ, Li H, Li WZ, Lin S, Dai AT, Cai XQ, Liu YY, Xu YN, Feng WB, Yang DH, Wang MW

EMDB-35707: 
Cryo-EM structure of GIPR splice variant 2 (SV2) in complex with Gs protein
Method: single particle / : Zhao FH, Hang KN, Zhou QT, Shao LJ, Li H, Li WZ, Lin S, Dai AT, Cai XQ, Liu YY, Xu YN, Feng WB, Yang DH, Wang MW

EMDB-40407: 
Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2)
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

EMDB-40408: 
Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 1)
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

EMDB-40409: 
Cryo-EM structure of a single loaded human UBA7-UBE2L6-ISG15 adenylate complex
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

EMDB-40782: 
Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex from a composite map
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

PDB-8se9: 
Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2)
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

PDB-8sea: 
Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 1)
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

PDB-8seb: 
Cryo-EM structure of a single loaded human UBA7-UBE2L6-ISG15 adenylate complex
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

PDB-8sv8: 
Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex from a composite map
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

EMDB-35254: 
ACE2-SIT1 complex bound with proline
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-35255: 
ACE2-B0AT1 complex bound with glutamine
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH
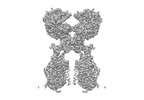
EMDB-35256: 
ACE2-B0AT1 complex bound with methionine
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-35260: 
Cryo-EM map of the ACE2-SIT1 complex bound with proline, focused refined on extracellular region
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-35261: 
cryo-EM map of the ACE2-SIT1 complex bound with proline, focused refined on transmembrane region
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-35262: 
cryo-EM map of the ACE2-B0AT1 complex bound with glutamine, focused refined on extracellular region
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-35265: 
cryo-EM map of the ACE2-B0AT1 complex bound with glutamine, focused refined on transmembrane region
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-35271: 
cryo-EM map of the ACE2-B0AT1 complex bound with methionine, focused refined on extracellular region
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-35273: 
cryo-EM map of the ACE2-B0AT1 complex bound with methionine, focused refined on transmembrane region
Method: single particle / : Li YN, Zhang YY, Shen YP, Yan RH

EMDB-36342: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist
Method: single particle / : He BB, Zhong YX, Guo Q, Tao YY

EMDB-36360: 
cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist
Method: single particle / : He BB, Zhong YX, Guo Q, Tao YY

EMDB-36361: 
Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist
Method: single particle / : He BB, Zhong YX, Guo Q, Tao YY

EMDB-34259: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv282
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34260: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv282 focused on RBD_XGv282 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q
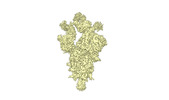
EMDB-34261: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv289
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34262: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv289 focused on RBD_XGv289 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q
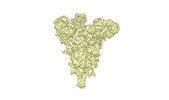
EMDB-34263: 
cryo-EM structure of Omicron BA.5 S protein in complex with S2L20
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34264: 
Cryo-EM map of Omicron BA.5 S protein in complex with S2L20 focused on NTD_S2L20 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q

EMDB-33633: 
Cryo-EM structure of C5a-bound C5aR1 in complex with Gi protein
Method: single particle / : Feng YY, Zhao C, Yan W, Shao ZH

EMDB-33634: 
Cryo-EM structure of C5a peptide-bound C5aR1 in complex with Gi protein
Method: single particle / : Feng YY, Zhao C, Yan W, Shao ZH

EMDB-33635: 
Cryo-EM structure of BM213-bound C5aR1 in complex with Gi protein
Method: single particle / : Feng YY, Zhao C, Yan W, Shao ZH

EMDB-33636: 
Cryo-EM structure of C089-bound C5aR1(I116A) mutant in complex with Gi protein
Method: single particle / : Feng YY, Zhao C, Yan W, Shao ZH

EMDB-32227: 
Cryo-EM structure of amyloid fibril formed by full-length human SOD1
Method: helical / : Wang LQ, Ma YY, Yuan HY, Zhao K, Zhang MY, Wang Q, Huang X, Xu WC, Chen J, Li D, Zhang DL, Zou LY, Yin P, Liu C, Liang Y

EMDB-30947: 
Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-30948: 
Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-30949: 
Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S
Method: single particle / : Guo YY, Zhang YY, Yan RH, Huang BD, Ye FF, Wu LS, Chi XM, Zhou Q

EMDB-31249: 
S protein of SARS-CoV-2 in complex with GW01
Method: single particle / : Shen YP, Zhang YY, Yan RH, Li YN, Zhou Q

EMDB-31250: 
Local map of S protein of SARS-CoV-2 in complex with GW01 Focused on RND-GW01 sub_complex
Method: single particle / : Shen YP, Zhang YY

EMDB-32920: 
S protein of SARS-CoV-2 in complex with 2G1
Method: single particle / : Guo YY, Zhang YY

EMDB-32921: 
S protein of SARS-CoV-2 in complex with 2G1 focused on RBD_2G1 sub-complex
Method: single particle / : Guo YY, Zhang YY

EMDB-30887: 
Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K
Method: helical / : Wang LQ, Zhao K, Yuan HY, Li XN, Dang HB, Ma YY, Wang Q, Wang C, Sun YP, Chen J, Li D, Zhang DL, Yin P, Liu C, Liang Y

EMDB-31146: 
Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading
Method: single particle / : Yan L, Yang YX

EMDB-31138: 
Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading
Method: single particle / : Yan LM, Yang YX, Li MY, Zhang Y, Zheng LT, Ge J, Huang YC, Liu ZY, Wang T, Gao S, Zhang R, Huang YY, Guddat LW, Gao Y, Rao ZH, Lou ZY
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
