-Search query
-Search result
Showing 1 - 50 of 56 items for (author: hall & dm)

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
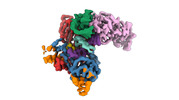
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-16512: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle
Method: single particle / : Bufton JC, Capin J, Boruku U, Garzoni F, Schaffitzel C, Berger I

EMDB-16522: 
Structure of ADDoCoV-ADAH11
Method: single particle / : Yadav KNS, Buzas D, Berger-Schaffitzel C, Berger I

PDB-8c9n: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle
Method: single particle / : Bufton JC, Capin J, Boruku U, Garzoni F, Schaffitzel C, Berger I
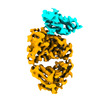
EMDB-16805: 
Cryo-EM structure of PcrV/Fab(30-B8)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

EMDB-16807: 
Cryo-EM structure of PcrV/Fab(11-E5)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

PDB-8cr9: 
Cryo-EM structure of PcrV/Fab(30-B8)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

PDB-8crb: 
Cryo-EM structure of PcrV/Fab(11-E5)
Method: single particle / : Yuan B, Simonis A, Marlovits TC

EMDB-16024: 
SARS-CoV-2 S protein in complex with pT1644 Fab
Method: single particle / : Stroeh L, Hansen G, Vollmer B, Krey T, Benecke T, Gruenewald K

EMDB-16026: 
SARS-CoV-2 S protein in complex with pT1696 Fab
Method: single particle / : Hansen G, Benecke T, Vollmer B, Gruenewald K, Krey T

EMDB-28825: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

EMDB-28827: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

PDB-8f2r: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

PDB-8f2u: 
Human CCC complex
Method: single particle / : Healy MD, McNally KE, Butkovic R, Chilton M, Kato K, Sacharz J, McConville C, Moody ERR, Shaw S, Planelles-Herrero VJ, Kadapalakere SY, Ross J, Borucu U, Palmer CS, Chen K, Croll TI, Hall RJ, Caruana NJ, Ghai R, Nguyen THD, Heesom KJ, Saitoh S, Berger I, Berger-Schaffitzel C, Williams TA, Stroud DA, Derivery E, Collins BM, Cullen PJ

EMDB-15606: 
CryoEM structure of the human Nucleophosmin 1 core
Method: single particle / : Valentin Gese G, Hallberg BM

PDB-8as5: 
CryoEM structure of the human Nucleophosmin 1 core
Method: single particle / : Valentin Gese G, Hallberg BM

EMDB-27415: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIa
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

EMDB-27416: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

EMDB-27417: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex III
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

EMDB-27420: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IV
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

EMDB-27423: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ia
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

EMDB-27427: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ib
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

PDB-8dfv: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIa
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

PDB-8dg5: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

PDB-8dg7: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex III
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

PDB-8dga: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IV
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

PDB-8dgi: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ia
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

PDB-8dgj: 
Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ib
Method: single particle / : Jouravleva K, Golovenko D, Demo G, Dutcher RC, Tanaka Hall TM, Zamore PD, Korostelev AA

EMDB-26847: 
Geometrically programmed DNA origami colloid for self-assembly of tubules: V-particle
Method: single particle / : Hayakawa D, Videbaek TE, Hall DM, Fang H, Sigl C, Feigl E, Dietz H, Fraden S, Hagan MF, Grason GM, Rogers WB

EMDB-26848: 
Geometrically programmed DNA origami colloid for self-assembly of tubules: X-particle
Method: single particle / : Hayakawa D, Videbaek TE, Hall DM, Fang H, Sigl C, Feigl E, Dietz H, Fraden S, Hagan MF, Grason GM, Rogers WB

EMDB-26849: 
Geometrically programmed DNA origami colloid for self-assembly of tubules: A-particle
Method: single particle / : Hayakawa D, Videbaek TE, Hall DM, Fang H, Sigl C, Feigl E, Dietz H, Fraden S, Hagan MF, Grason GM, Rogers WB

EMDB-26850: 
Geometrically programmed DNA origami colloid for self-assembly of tubules: B-particle
Method: single particle / : Hayakawa D, Videbaek TE, Hall DM, Fang H, Sigl C, Feigl E, Dietz H, Fraden S, Hagan MF, Grason GM, Rogers WB

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-7uhb: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-7uhc: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-13218: 
Progressive supranuclear palsy tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW

EMDB-13219: 
Globular glial tauopathy type 1 tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW

EMDB-13220: 
Globular glial tauopathy type 2 tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW

EMDB-13221: 
Globular glial tauopathy type 3 tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW

EMDB-13223: 
Limbic-predominant neuronal inclusion body 4R tauopathy type 1a tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW

EMDB-13224: 
Limbic-predominant neuronal inclusion body 4R tauopathy type 1b tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW

EMDB-13225: 
Limbic-predominant neuronal inclusion body 4R tauopathy type 2 tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW

EMDB-13226: 
Argyrophilic grain disease type 1 tau filament
Method: helical / : Shi Y, Zhang W, Yang Y, Murzin AG, Falcon B, Kotecha A, van Beers M, Tarutani A, Kametani F, Garringer HJ, Vidal R, Hallinan GI, Lashley T, Saito Y, Murayama S, Yoshida M, Tanaka H, Kakita A, Ikeuchi T, Robinson AC, Mann DMA, Kovacs GG, Revesz T, Ghetti B, Hasegawa M, Goedert M, Scheres SHW
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
