-Search query
-Search result
Showing 1 - 50 of 370 items for (author: egan & es)

EMDB-19576: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-19582: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : LM32Cs3H1 sKO STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8rxh: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8rxx: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : LM32Cs3H1 sKO STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-18639: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)
Method: single particle / : Ren J, Stuart DI, Duyvesteyn HME

EMDB-18649: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
Method: single particle / : Ren J, Duyvesteyn HME, Stuart DI

EMDB-19002: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8qsq: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)
Method: single particle / : Ren J, Stuart DI, Duyvesteyn HME

PDB-8qtd: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
Method: single particle / : Ren J, Duyvesteyn HME, Stuart DI

PDB-8r8k: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-17216: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8ovj: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17863: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod
Method: helical / : Hospenthal M, Zyla D, Glockshuber R, Waksman G

EMDB-17878: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod
Method: helical / : Zyla D, Hospenthal M, Glockshuber R, Waksman G

PDB-8psv: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod
Method: helical / : Hospenthal M, Zyla D, Glockshuber R, Waksman G

PDB-8ptu: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod
Method: helical / : Zyla D, Hospenthal M, Glockshuber R, Waksman G

EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
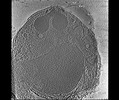
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-43271: 
Cryo-EM structure of the Helicobacter pylori VacA hexamer that was detergent solubilized from membrane, C6 symmetry applied
Method: single particle / : Connolly SM, Ohi MD

EMDB-43272: 
Cryo-EM structure of Helicobacter pylori VacA hexamer that was detergent solubilized form membrane, no symmetry applied
Method: single particle / : Connolly SM, Ohi MD

EMDB-42124: 
Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab
Method: single particle / : Li F, Bailis JM

PDB-8ucd: 
Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab
Method: single particle / : Li F, Bailis JM, Zhang H

EMDB-15272: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT
Method: single particle / : Rajan KS, Yonath A, Bashan A

PDB-8a98: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT
Method: single particle / : Rajan KS, Yonath A, Bashan A

EMDB-28125: 
Plasmodium falciparum merozoites apical 2-ring units
Method: subtomogram averaging / : Sun SY, Pintilie GD
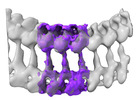
EMDB-28126: 
Toxoplasma apical rings
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-40272: 
BtCoV-422 in complex with neutralizing antibody JC57-11
Method: single particle / : McFadden E, McLellan JS

EMDB-40306: 
BtCoV-422 spike in complex with JC57-11 Fab, global map
Method: single particle / : McFadden E, McLellan JS

EMDB-40310: 
BtCoV-422 spike in complex with JC57-11 Fab, local map
Method: single particle / : McFadden E, McLellan JS

PDB-8sak: 
BtCoV-422 in complex with neutralizing antibody JC57-11
Method: single particle / : McFadden E, McLellan JS

EMDB-18301: 
In-tissue cryo electron tomograms of App^NL-G-F amyloid plaques
Method: electron tomography / : Leistner C, Wilkinson M, Burgess A, Lovatt M, Goodbody S, Xu Y, Deuchars S, Radford SE, Ranson NA, Frank RAW

EMDB-29280: 
Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA
Method: single particle / : Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin C, Buyukyoruk M, Neselu K, Eng ET, Lander GC, Wiedenheft B

PDB-8flj: 
Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA
Method: single particle / : Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin C, Buyukyoruk M, Neselu K, Eng ET, Lander GC, Wiedenheft B

EMDB-27847: 
Mouse norovirus strain CR6, attenuated
Method: single particle / : Smith TJ, Sherman M

EMDB-27849: 
Mouse norovirus strain CR6 at pH 5.0
Method: single particle / : Smith TJ

EMDB-27692: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-27693: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (global refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-8dt8: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-27205: 
Closed state of SARS-CoV-2 BA.2 variant spike protein
Method: single particle / : Zhang J, Tang WC, Gao HL, Shi W, Peng HQ, Volloch SR, Xiao TS, Chen B

EMDB-27206: 
One RBD-up state of SARS-CoV-2 BA.2 variant spike protein
Method: single particle / : Zhang J, Tang WC, Gao HL, Shi W, Peng HQ, Volloch SR, Xiao TS, Chen B

EMDB-27207: 
Middle state of SARS-CoV-2 BA.2 variant spike protein
Method: single particle / : Zhang J, Tang WC, Gao HL, Shi W, Peng HQ, Volloch SR, Xiao TS, Chen B

PDB-8d55: 
Closed state of SARS-CoV-2 BA.2 variant spike protein
Method: single particle / : Zhang J, Tang WC, Gao HL, Shi W, Peng HQ, Volloch SR, Xiao TS, Chen B

PDB-8d56: 
One RBD-up state of SARS-CoV-2 BA.2 variant spike protein
Method: single particle / : Zhang J, Tang WC, Gao HL, Shi W, Peng HQ, Volloch SR, Xiao TS, Chen B

PDB-8d5a: 
Middle state of SARS-CoV-2 BA.2 variant spike protein
Method: single particle / : Zhang J, Tang WC, Gao HL, Shi W, Peng HQ, Volloch SR, Xiao TS, Chen B

EMDB-16018: 
Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure
Method: helical / : Wilkinson M, Leistner C, Burgess A, Goodfellow S, Deuchars S, Ranson NA, Radford SE, Frank RAW

EMDB-16019: 
Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice)
Method: helical / : Wilkinson M, Leistner C, Burgess A, Goodfellow S, Deuchars S, Ranson NA, Radford SE, Frank RAW

PDB-8bfa: 
Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure
Method: helical / : Wilkinson M, Leistner C, Burgess A, Goodfellow S, Deuchars S, Ranson NA, Radford SE, Frank RAW
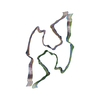
PDB-8bfb: 
Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice)
Method: helical / : Wilkinson M, Leistner C, Burgess A, Goodfellow S, Deuchars S, Ranson NA, Radford SE, Frank RAW

EMDB-27319: 
Mouse Norovirus strain WU23
Method: single particle / : Smith TJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
