[English] 日本語
 Yorodumi
Yorodumi- EMDB-38829: 1up-1 conformation of HKU1-B S protein after incubation of the re... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1up-1 conformation of HKU1-B S protein after incubation of the receptor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HCoV-HKU1 /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationendocytosis involved in viral entry into host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / host cell plasma membrane / virion membrane / viral envelope / host cell plasma membrane / virion membrane /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |  Human coronavirus HKU1 (isolate N2) Human coronavirus HKU1 (isolate N2) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Xia LY / Zhang YY / Zhou Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2. Authors: Lingyun Xia / Yuanyuan Zhang / Qiang Zhou /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38829.map.gz emd_38829.map.gz | 230.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38829-v30.xml emd-38829-v30.xml emd-38829.xml emd-38829.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38829_fsc.xml emd_38829_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_38829.png emd_38829.png | 88.5 KB | ||
| Filedesc metadata |  emd-38829.cif.gz emd-38829.cif.gz | 6.2 KB | ||
| Others |  emd_38829_half_map_1.map.gz emd_38829_half_map_1.map.gz emd_38829_half_map_2.map.gz emd_38829_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38829 http://ftp.pdbj.org/pub/emdb/structures/EMD-38829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38829 | HTTPS FTP |
-Related structure data
| Related structure data |  8y1aMC  8y19C  8y1bC  8y1cC  8y1dC  8y1eC  8y1fC  8y1gC  8y1hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38829.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38829.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38829_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
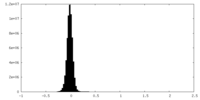
| Density Histograms |
-Half map: #1
| File | emd_38829_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
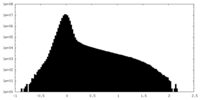
| Density Histograms |
- Sample components
Sample components
-Entire : 1up-1 conformation of HKU1-B S protein after incubation of the re...
| Entire | Name: 1up-1 conformation of HKU1-B S protein after incubation of the receptor |
|---|---|
| Components |
|
-Supramolecule #1: 1up-1 conformation of HKU1-B S protein after incubation of the re...
| Supramolecule | Name: 1up-1 conformation of HKU1-B S protein after incubation of the receptor type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human coronavirus HKU1 (isolate N2) Human coronavirus HKU1 (isolate N2) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human coronavirus HKU1 (isolate N2) Human coronavirus HKU1 (isolate N2) |
| Molecular weight | Theoretical: 143.475344 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFLIIFILPT TLAVIGDFNC TNSFINDYNK TIPRISEDVV DVSLGLGTYY VLNRVYLNTT LLFTGYFPKS GANFRDLALK GSKYLSTLW YKPPFLSDFN NGIFSKVKNT KLYVNNTLYS EFSTIVIGSV FVNTSYTIVV QPHNGILEIT ACQYTMCEYP H TVCKSKGS ...String: MFLIIFILPT TLAVIGDFNC TNSFINDYNK TIPRISEDVV DVSLGLGTYY VLNRVYLNTT LLFTGYFPKS GANFRDLALK GSKYLSTLW YKPPFLSDFN NGIFSKVKNT KLYVNNTLYS EFSTIVIGSV FVNTSYTIVV QPHNGILEIT ACQYTMCEYP H TVCKSKGS IRNESWHIDS SEPLCLFKKN FTYNVSADWL YFHFYQERGV FYAYYADVGM PTTFLFSLYL GTILSHYYVM PL TCKAISS NTDNETLEYW VTPLSRRQYL LNFDEHGVIT NAVDCSSSFL SEIQCKTQSF APNTGVYDLS GFTVKPVATV YRR IPNLPD CDIDNWLNNV SVPSPLNWER RIFSNCNFNL STLLRLVHVD SFSCNNLDKS KIFGSCFNSI TVDKFAIPNR RRDD LQLGS SGFLQSSNYK IDISSSSCQL YYSLPLVNVT INNFNPSSWN RRYGFGSFNV SSYDVVYSDH CFSVNSDFCP CADPS VVNS CVKSKPLSAI CPAGTKYRHC DLDTTLYVNN WCRCSCLPDP ISTYSPNTCP QKKVVVGIGE HCPGLGINEE KCGTQL NHS SCSCSPDAFL GWSFDSCISN NRCNIFSNFI FNGINSGTTC SNDLLYSNTE VSTGVCVNYD LYGITGQGIF KEVSAAY YN NWQNLLYDSN GNIIGFKDFL TNKTYTILPC YSGRVSAAFY QNSSSPALLY RNLKCSYVLN NISFISQPFY FDSYLGCV L NAVNLTSYSV SSCDLRMGSG FCIDYALPSS GSASRGISSP YRFVTFEPFN VSFVNDSVET VGGLFEIQIP TNFTIAGHE EFIQTSSPKV TIDCSAFVCS NYAACHDLLS EYGTFCDNIN SILNEVNDLL DITQLQVANA LMQGVTLSSN LNTNLHSDVD NIDFKSLLG CLGSQCGSSS RSLLEDLLFN KVKLSDVGFV EAYNNCTGGS EIRDLLCVQS FNGIKVLPPI LSETQISGYT T AATVAAMF PPWSAAAGVP FSLNVQYRIN GLGVTMDVLN KNQKLIANAF NKALLSIQNG FTATNSALAK IQSVVNANAQ AL NSLLQQL FNKFGAISSS LQEILSRLDP PEAQVQIDRL INGRLTALNA YVSQQLSDIT LIKAGASRAI EKVNECVKSQ SPR INFCGN GNHILSLVQN APYGLLFIHF SYKPTSFKTV LVSPGLCLSG DRGIAPKQGY FIKQNDSWMF TGSSYYYPEP ISDK NVVFM NSCSVNFTKA PFIYLNNSIP NLSDFEAEFS LWFKNHTSIA PNLTFNSHIN ATFLDLYYEM NVIQESIKSL NSSFI NLKE IGTYEM UniProtKB:  Spike glycoprotein Spike glycoprotein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 51 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X