+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8t79 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
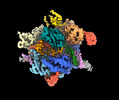
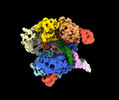
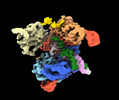
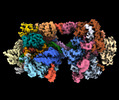
| Title | SpRY-Cas9:gRNA complex bound to non-target DNA with 10 bp R-loop | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  IMMUNE SYSTEM / SpRY-Cas9 / IMMUNE SYSTEM / SpRY-Cas9 /  CRISPR / CRISPR /  Cas9 / Cas9 /  R-loop R-loop | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / 3'-5' exonuclease activity / DNA endonuclease activity / defense response to virus /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  DNA binding / DNA binding /  RNA binding / RNA binding /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria)  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.04 Å cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Hibshman, G.N. / Bravo, J.P.K. / Taylor, D.W. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9. Authors: Grace N Hibshman / Jack P K Bravo / Matthew M Hooper / Tyler L Dangerfield / Hongshan Zhang / Ilya J Finkelstein / Kenneth A Johnson / David W Taylor /   Abstract: CRISPR-Cas9 is a powerful tool for genome editing, but the strict requirement for an NGG protospacer-adjacent motif (PAM) sequence immediately next to the DNA target limits the number of editable ...CRISPR-Cas9 is a powerful tool for genome editing, but the strict requirement for an NGG protospacer-adjacent motif (PAM) sequence immediately next to the DNA target limits the number of editable genes. Recently developed Cas9 variants have been engineered with relaxed PAM requirements, including SpG-Cas9 (SpG) and the nearly PAM-less SpRY-Cas9 (SpRY). However, the molecular mechanisms of how SpRY recognizes all potential PAM sequences remains unclear. Here, we combine structural and biochemical approaches to determine how SpRY interrogates DNA and recognizes target sites. Divergent PAM sequences can be accommodated through conformational flexibility within the PAM-interacting region, which facilitates tight binding to off-target DNA sequences. Nuclease activation occurs ~1000-fold slower than for Streptococcus pyogenes Cas9, enabling us to directly visualize multiple on-pathway intermediate states. Experiments with SpG position it as an intermediate enzyme between Cas9 and SpRY. Our findings shed light on the molecular mechanisms of PAMless genome editing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8t79.cif.gz 8t79.cif.gz | 317.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8t79.ent.gz pdb8t79.ent.gz | 243.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8t79.json.gz 8t79.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/t7/8t79 https://data.pdbj.org/pub/pdb/validation_reports/t7/8t79 ftp://data.pdbj.org/pub/pdb/validation_reports/t7/8t79 ftp://data.pdbj.org/pub/pdb/validation_reports/t7/8t79 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  41088MC  8spqC  8sqhC  8srsC  8t6oC  8t6pC  8t6sC  8t6tC  8t6xC  8t6yC  8t76C  8t77C  8t78C  8t7sC  8tzzC  8u3yC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 158676.031 Da / Num. of mol.: 1 Mutation: A61R, L1111R, D1135L, S1136W, G1218K, E1219Q, N1317R, A1322R, R1333P, R1335Q, T1337R Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Streptococcus pyogenes (bacteria) / Gene: cas9, csn1, SPy_1046 / Production host: Streptococcus pyogenes (bacteria) / Gene: cas9, csn1, SPy_1046 / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: Q99ZW2,  Hydrolases; Acting on ester bonds Hydrolases; Acting on ester bonds |
|---|---|
| #2: RNA chain |  Guide RNA Guide RNAMass: 28724.102 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| #3: DNA chain | Mass: 6032.886 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| #4: DNA chain | Mass: 2813.875 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Ternary complex of SpRY-Cas9 with gRNA and non-target DNA with 10 bp R-loop Type: COMPLEX / Entity ID: all / Source: MULTIPLE SOURCES |
|---|---|
| Molecular weight | Value: 0.21154 MDa / Experimental value: YES |
| Source (natural) | Organism:   Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Source (recombinant) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 1500 nm Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 1500 nm |
| Image recording | Electron dose: 80 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
3D reconstruction | Resolution: 3.04 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 123454 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj































































