[English] 日本語
 Yorodumi
Yorodumi- PDB-8ja2: ASFV Topoisomerase ATPase domain in complex with AMP-PNP and Mg2+ -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8ja2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
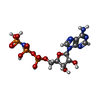
| Title | ASFV Topoisomerase ATPase domain in complex with AMP-PNP and Mg2+ | |||||||||
 Components Components | DNA topoisomerase 2 Topoisomerase Topoisomerase | |||||||||
 Keywords Keywords |  ISOMERASE / GHKL NUCLEOTIDE-BINDING FOLD / ISOMERASE / GHKL NUCLEOTIDE-BINDING FOLD /  Topoisomerase / AMP-PNP Topoisomerase / AMP-PNP | |||||||||
| Function / homology |  Function and homology information Function and homology informationsister chromatid segregation / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity /  DNA topoisomerase (ATP-hydrolysing) / DNA topological change / host cell cytoplasm / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / host cell cytoplasm /  DNA binding / DNA binding /  ATP binding / ATP binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |    African swine fever virus African swine fever virus | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.73 Å MOLECULAR REPLACEMENT / Resolution: 1.73 Å | |||||||||
 Authors Authors | Pang, A.H. / Chang, C.-W. / Tsai, M.-D. | |||||||||
| Funding support |  Taiwan, 2items Taiwan, 2items
| |||||||||
 Citation Citation |  Journal: Commun Chem / Year: 2024 Journal: Commun Chem / Year: 2024Title: A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase. Authors: Chiung-Wen Mary Chang / Shun-Chang Wang / Chun-Hsiung Wang / Allan H Pang / Cheng-Han Yang / Yao-Kai Chang / Wen-Jin Wu / Ming-Daw Tsai /   Abstract: The theories for substrate recognition in enzyme catalysis have evolved from lock-key to induced fit, then conformational selection, and conformational selection followed by induced fit. However, the ...The theories for substrate recognition in enzyme catalysis have evolved from lock-key to induced fit, then conformational selection, and conformational selection followed by induced fit. However, the prevalence and consensus of these theories require further examination. Here we use cryogenic electron microscopy and African swine fever virus type 2 topoisomerase (AsfvTop2) to demonstrate substrate binding theories in a joint and ordered manner: catalytic selection by the enzyme, conformational selection by the substrates, then induced fit. The apo-AsfvTop2 pre-exists in six conformers that comply with the two-gate mechanism directing DNA passage and release in the Top2 catalytic cycle. The structures of AsfvTop2-DNA-inhibitor complexes show that substantial induced-fit changes occur locally from the closed apo-conformer that however is too far-fetched for the open apo-conformer. Furthermore, the ATPase domain of AsfvTop2 in the MgAMP-PNP-bound crystal structures coexist in reduced and oxidized forms involving a disulfide bond, which can regulate the AsfvTop2 function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8ja2.cif.gz 8ja2.cif.gz | 98 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8ja2.ent.gz pdb8ja2.ent.gz | 70.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8ja2.json.gz 8ja2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ja/8ja2 https://data.pdbj.org/pub/pdb/validation_reports/ja/8ja2 ftp://data.pdbj.org/pub/pdb/validation_reports/ja/8ja2 ftp://data.pdbj.org/pub/pdb/validation_reports/ja/8ja2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8j87C  8j88C  8j89C  8j8aC  8j8bC  8j8cC  8j9vC  8j9wC  8j9xC  8ja1C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
| ||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein |  Topoisomerase TopoisomeraseMass: 45463.555 Da / Num. of mol.: 1 / Fragment: ATPase domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)    African swine fever virus / Gene: P1192R / Production host: African swine fever virus / Gene: P1192R / Production host:   Escherichia coli (E. coli) / References: UniProt: A0A0A1E3Q0 Escherichia coli (E. coli) / References: UniProt: A0A0A1E3Q0 |
|---|---|
| #2: Chemical | ChemComp-MG / |
| #3: Chemical | ChemComp-ANP / |
| #4: Water | ChemComp-HOH /  Water Water |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.49 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7.5 / Details: PEG 3350, Tris, LiSO4 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSRRC NSRRC  / Beamline: TPS 05A / Wavelength: 0.9998 Å / Beamline: TPS 05A / Wavelength: 0.9998 Å |
| Detector | Type: RAYONIX MX300-HS / Detector: CCD / Date: Feb 28, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9998 Å / Relative weight: 1 : 0.9998 Å / Relative weight: 1 |
| Reflection | Resolution: 1.73→43.21 Å / Num. obs: 48808 / % possible obs: 99 % / Redundancy: 8.6 % / Biso Wilson estimate: 17.81 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.069 / Net I/σ(I): 18.6 |
| Reflection shell | Resolution: 1.73→1.82 Å / Redundancy: 8.4 % / Rmerge(I) obs: 0.404 / Num. unique obs: 6982 / CC1/2: 0.934 / % possible all: 98.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 1.73→43.17 Å / SU ML: 0.2026 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 20.5368 MOLECULAR REPLACEMENT / Resolution: 1.73→43.17 Å / SU ML: 0.2026 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 20.5368 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20.35 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.73→43.17 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj