[English] 日本語
 Yorodumi
Yorodumi- EMDB-36473: Cryo-EM structure of the full-length African swine fever virus to... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 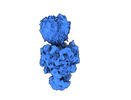 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the full-length African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Topoisomerase / Topoisomerase /  ASFV / ASFV /  inhibitor / inhibitor /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Biological species |    African swine fever virus African swine fever virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.68 Å cryo EM / Resolution: 3.68 Å | |||||||||
 Authors Authors | Chang CW / Tsai MD | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Chem / Year: 2024 Journal: Commun Chem / Year: 2024Title: A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase. Authors: Chiung-Wen Mary Chang / Shun-Chang Wang / Chun-Hsiung Wang / Allan H Pang / Cheng-Han Yang / Yao-Kai Chang / Wen-Jin Wu / Ming-Daw Tsai /   Abstract: The theories for substrate recognition in enzyme catalysis have evolved from lock-key to induced fit, then conformational selection, and conformational selection followed by induced fit. However, the ...The theories for substrate recognition in enzyme catalysis have evolved from lock-key to induced fit, then conformational selection, and conformational selection followed by induced fit. However, the prevalence and consensus of these theories require further examination. Here we use cryogenic electron microscopy and African swine fever virus type 2 topoisomerase (AsfvTop2) to demonstrate substrate binding theories in a joint and ordered manner: catalytic selection by the enzyme, conformational selection by the substrates, then induced fit. The apo-AsfvTop2 pre-exists in six conformers that comply with the two-gate mechanism directing DNA passage and release in the Top2 catalytic cycle. The structures of AsfvTop2-DNA-inhibitor complexes show that substantial induced-fit changes occur locally from the closed apo-conformer that however is too far-fetched for the open apo-conformer. Furthermore, the ATPase domain of AsfvTop2 in the MgAMP-PNP-bound crystal structures coexist in reduced and oxidized forms involving a disulfide bond, which can regulate the AsfvTop2 function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36473.map.gz emd_36473.map.gz | 152.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36473-v30.xml emd-36473-v30.xml emd-36473.xml emd-36473.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36473_fsc.xml emd_36473_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_36473.png emd_36473.png | 46.3 KB | ||
| Filedesc metadata |  emd-36473.cif.gz emd-36473.cif.gz | 5.3 KB | ||
| Others |  emd_36473_half_map_1.map.gz emd_36473_half_map_1.map.gz emd_36473_half_map_2.map.gz emd_36473_half_map_2.map.gz | 285.5 MB 285.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36473 http://ftp.pdbj.org/pub/emdb/structures/EMD-36473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36473 | HTTPS FTP |
-Related structure data
| Related structure data |  8j87C  8j88C  8j89C  8j8aC  8j8bC  8j8cC  8j9vC  8j9wC  8j9xC  8ja1C  8ja2C C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36473.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36473.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36473_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36473_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ASFV Topoisomerase 2 complexed with Cut02aDNA and etoposide
| Entire | Name: ASFV Topoisomerase 2 complexed with Cut02aDNA and etoposide |
|---|---|
| Components |
|
-Supramolecule #1: ASFV Topoisomerase 2 complexed with Cut02aDNA and etoposide
| Supramolecule | Name: ASFV Topoisomerase 2 complexed with Cut02aDNA and etoposide type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:    African swine fever virus African swine fever virus |
-Macromolecule #1: African swine fever virus type 2 topoisomerase
| Macromolecule | Name: African swine fever virus type 2 topoisomerase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    African swine fever virus African swine fever virus |
| Recombinant expression | Organism:   Saccharomyces (fungus) Saccharomyces (fungus) |
| Sequence | String: MEAFEISDFK EHAKKKSMWA GALNKVTISG LMGVFTEDED LMALPIHRDH CPALLKIFDE LIVNATDHE RACHSKTKKV TYIKISFDKG VFSCENDGPG IPIAKHEQAS LIAKRDVYVP E VASCFFLA GTNINKAKDC IKGGTNGVGL KLAMVHSQWA ILTTADGAQK ...String: MEAFEISDFK EHAKKKSMWA GALNKVTISG LMGVFTEDED LMALPIHRDH CPALLKIFDE LIVNATDHE RACHSKTKKV TYIKISFDKG VFSCENDGPG IPIAKHEQAS LIAKRDVYVP E VASCFFLA GTNINKAKDC IKGGTNGVGL KLAMVHSQWA ILTTADGAQK YVQQINQRLD II EPPTITP SREMFTRIEL MPVYQELGYA EPLSETEQAD LSAWIYLRAC QCAAYVGKGT TIY YNDKPC RTGSVMALAK MYTLLSAPNS TIHTATIKAD AKPYSLHPLQ VAAVVSPKFK KFEH VSIIN GVNCVKGEHV TFLKKTINEM VIKKFQQTIK DKNRKTTLRD SCSNIFVVIV GSIPG IEWT GQRKDELSIA ENVFKTHYSI PSSFLTSMTR SIVDILLQSI SKKDNHKQVD VDKYTR ARN AGGKRAQDCM LLAAEGDSAL SLLRTGLTLG KSNPSGPSFD FCGMISLGGV IMNACKK VT NITTDSGETI MVRNEQLTNN KVLQGIVQVL GLDFNCHYKT QEERAKLRYG CIVACVDQ D LDGCGKILGL LLAYFHLFWP QLIIHGFVKR LLTPLIRVYE KGKTMPVEFY YEQEFDAWA KKQTSLVNHT VKYYKGLAAH DTHEVKSMFK HFDNMVYTFT LDDSAKELFH IYFGGESELR KRELCTGVV PLTETQTQSI HSVRRIPCSL HLQVDTKAYK LDAIERQIPN FLDGMTRARR K ILAGGVKC FASNNRERKV FQFGGYVADH MFYHHGDMSL NTSIIKAAQY YPGSSHLYPV FI GIGSFGS RHLGGKDAGS PRYISVQLAS EFIKTMFPAE DSWLLPYVFE DGQRAEPEYY VPV LPLAIM EYGANPSEGW KYTTWARQLE DILALVRAYV DKDNPKHELL HYAIKHKITI LPLR PSNYN FKGHLKRFGQ YYYSYGTYDI SEQRNIITIT ELPLRVPTVA YIESIKKSSN RMTFI EEII DYSSSETIEI LVKLKPNSLN RIVEEFKETE EQDSIENFLR LRNCLHSHLN FVKPKG GII EFNSYYEILY AWLPYRRELY QKRLMREHAV LKLRIIMETA IVRYINESAE LNLSHYE DE KEASRILSEH GFPPLNHTLI ISPEFASIEE LNQKALQGCY TYILSLQARE LLIAAKTR R VEKIKKMQAR LDKVEQLLQE SPFPGASVWL EEIDAVEKAI IKGRNTQWKF H |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11786 / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X


















