[English] 日本語
 Yorodumi
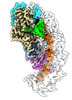
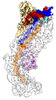
Yorodumi- PDB-8g9t: Exploiting Activation and Inactivation Mechanisms in Type I-C CRI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8g9t | ||||||
|---|---|---|---|---|---|---|---|
| Title | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/RNA /  CRISPR / type I-C / cascade / CRISPR / type I-C / cascade /  anti-CRISPR / HYDROLASE-RNA complex anti-CRISPR / HYDROLASE-RNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements /  endonuclease activity / defense response to virus / endonuclease activity / defense response to virus /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  RNA binding RNA bindingSimilarity search - Function | ||||||
| Biological species |  Rhodobacter phage RcNL1 (virus) Rhodobacter phage RcNL1 (virus)  Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | ||||||
 Authors Authors | Hu, C. / Nam, K.H. / Ke, A. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications. Authors: Chunyi Hu / Mason T Myers / Xufei Zhou / Zhonggang Hou / Macy L Lozen / Ki Hyun Nam / Yan Zhang / Ailong Ke /    Abstract: Type I CRISPR-Cas systems utilize the RNA-guided Cascade complex to identify matching DNA targets and the nuclease-helicase Cas3 to degrade them. Among the seven subtypes, type I-C is compact in size ...Type I CRISPR-Cas systems utilize the RNA-guided Cascade complex to identify matching DNA targets and the nuclease-helicase Cas3 to degrade them. Among the seven subtypes, type I-C is compact in size and highly active in creating large-sized genome deletions in human cells. Here, we use four cryoelectron microscopy snapshots to define its RNA-guided DNA binding and cleavage mechanisms in high resolution. The non-target DNA strand (NTS) is accommodated by I-C Cascade in a continuous binding groove along the juxtaposed Cas11 subunits. Binding of Cas3 further traps a flexible bulge in NTS, enabling NTS nicking. We identified two anti-CRISPR proteins AcrIC8 and AcrIC9 that strongly inhibit Neisseria lactamica I-C function. Structural analysis showed that AcrIC8 inhibits PAM recognition through allosteric inhibition, whereas AcrIC9 achieves so through direct competition. Both Acrs potently inhibit I-C-mediated genome editing and transcriptional modulation in human cells, providing the first off-switches for type I CRISPR eukaryotic genome engineering. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8g9t.cif.gz 8g9t.cif.gz | 584.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8g9t.ent.gz pdb8g9t.ent.gz | 480.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8g9t.json.gz 8g9t.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g9/8g9t https://data.pdbj.org/pub/pdb/validation_reports/g9/8g9t ftp://data.pdbj.org/pub/pdb/validation_reports/g9/8g9t ftp://data.pdbj.org/pub/pdb/validation_reports/g9/8g9t | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  29878MC  8g9sC  8g9uC  8gafC  8gamC  8ganC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 5 types, 14 molecules ABCDEFHMGIJLKN
| #1: Protein | Mass: 8247.822 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Rhodobacter phage RcNL1 (virus) / Gene: RHZG_00032 / Production host: Rhodobacter phage RcNL1 (virus) / Gene: RHZG_00032 / Production host:   Escherichia coli BL21 (bacteria) / References: UniProt: I3UM23 Escherichia coli BL21 (bacteria) / References: UniProt: I3UM23 | ||||||
|---|---|---|---|---|---|---|---|
| #2: Protein | Mass: 32208.111 Da / Num. of mol.: 7 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Neisseria lactamica (bacteria) / Gene: NCTC10618_01084 / Production host: Neisseria lactamica (bacteria) / Gene: NCTC10618_01084 / Production host:   Escherichia coli (E. coli) / References: UniProt: A0A378VEU0 Escherichia coli (E. coli) / References: UniProt: A0A378VEU0#3: Protein | Mass: 14245.184 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Neisseria lactamica (bacteria) / Gene: NCTC10618_01085 / Production host: Neisseria lactamica (bacteria) / Gene: NCTC10618_01085 / Production host:   Escherichia coli (E. coli) / References: UniProt: A0A378VF47 Escherichia coli (E. coli) / References: UniProt: A0A378VF47#4: Protein | |  Roberval (Air Saguenay) Water Aerodrome Roberval (Air Saguenay) Water AerodromeMass: 64860.832 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Neisseria lactamica (bacteria) / Gene: NCTC10618_01085 / Production host: Neisseria lactamica (bacteria) / Gene: NCTC10618_01085 / Production host:   Escherichia coli (E. coli) / References: UniProt: A0A378VF47 Escherichia coli (E. coli) / References: UniProt: A0A378VF47#5: Protein | | Mass: 23870.451 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Neisseria lactamica (bacteria) / Gene: cas5d / Production host: Neisseria lactamica (bacteria) / Gene: cas5d / Production host:   Escherichia coli (E. coli) / References: UniProt: D0W8X4 Escherichia coli (E. coli) / References: UniProt: D0W8X4 |
-RNA chain , 1 types, 1 molecules O
| #6: RNA chain | Mass: 13870.306 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Binary complex of AcrIC9 with crRNA bound type I-C Cascade Type: COMPLEX / Entity ID: all / Source: MULTIPLE SOURCES | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.4 MDa / Experimental value: YES | |||||||||||||||
| Source (natural) |
| |||||||||||||||
| Source (recombinant) |
| |||||||||||||||
| Buffer solution | pH: 7.5 / Details: 25mM Tris pH 7.5, 150mM NaCl | |||||||||||||||
| Buffer component | Conc.: 150 mM / Name: sodium chloride / Formula: NaCl / Formula: NaCl Sodium chloride Sodium chloride | |||||||||||||||
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | |||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: Quantifoil R1.2/1.3 | |||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 278 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TALOS ARCTICA |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: DIFFRACTION / Nominal magnification: 67000 X / Nominal defocus max: 3500 nm / Nominal defocus min: 2500 nm / Calibrated defocus min: 1500 nm / Calibrated defocus max: 3000 nm / Cs / Nominal magnification: 67000 X / Nominal defocus max: 3500 nm / Nominal defocus min: 2500 nm / Calibrated defocus min: 1500 nm / Calibrated defocus max: 3000 nm / Cs : 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE : 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Temperature (max): 100 K / Temperature (min): 70 K |
| Image recording | Average exposure time: 2.5 sec. / Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of grids imaged: 1200 / Num. of real images: 1200 |
| EM imaging optics | Energyfilter name : GIF Bioquantum : GIF Bioquantum |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 200000 / Num. of class averages: 6 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj





























