[English] 日本語
 Yorodumi
Yorodumi- EMDB-29896: Exploiting Activation and Inactivation Mechanisms in Type I-C CRI... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
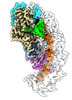
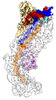
| Title | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  CRISPR / type I-C / cascade / CRISPR / type I-C / cascade /  anti-CRISPR / HYDROLASE-RNA complex anti-CRISPR / HYDROLASE-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements /  endonuclease activity / defense response to virus / endonuclease activity / defense response to virus /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  RNA binding RNA bindingSimilarity search - Function | |||||||||
| Biological species |   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.64 Å cryo EM / Resolution: 3.64 Å | |||||||||
 Authors Authors | Hu C / Nam KH / Ke A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications. Authors: Chunyi Hu / Mason T Myers / Xufei Zhou / Zhonggang Hou / Macy L Lozen / Ki Hyun Nam / Yan Zhang / Ailong Ke /    Abstract: Type I CRISPR-Cas systems utilize the RNA-guided Cascade complex to identify matching DNA targets and the nuclease-helicase Cas3 to degrade them. Among the seven subtypes, type I-C is compact in size ...Type I CRISPR-Cas systems utilize the RNA-guided Cascade complex to identify matching DNA targets and the nuclease-helicase Cas3 to degrade them. Among the seven subtypes, type I-C is compact in size and highly active in creating large-sized genome deletions in human cells. Here, we use four cryoelectron microscopy snapshots to define its RNA-guided DNA binding and cleavage mechanisms in high resolution. The non-target DNA strand (NTS) is accommodated by I-C Cascade in a continuous binding groove along the juxtaposed Cas11 subunits. Binding of Cas3 further traps a flexible bulge in NTS, enabling NTS nicking. We identified two anti-CRISPR proteins AcrIC8 and AcrIC9 that strongly inhibit Neisseria lactamica I-C function. Structural analysis showed that AcrIC8 inhibits PAM recognition through allosteric inhibition, whereas AcrIC9 achieves so through direct competition. Both Acrs potently inhibit I-C-mediated genome editing and transcriptional modulation in human cells, providing the first off-switches for type I CRISPR eukaryotic genome engineering. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29896.map.gz emd_29896.map.gz | 79.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29896-v30.xml emd-29896-v30.xml emd-29896.xml emd-29896.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29896.png emd_29896.png | 110.6 KB | ||
| Filedesc metadata |  emd-29896.cif.gz emd-29896.cif.gz | 6.7 KB | ||
| Others |  emd_29896_half_map_1.map.gz emd_29896_half_map_1.map.gz emd_29896_half_map_2.map.gz emd_29896_half_map_2.map.gz | 79.1 MB 79.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29896 http://ftp.pdbj.org/pub/emdb/structures/EMD-29896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29896 | HTTPS FTP |
-Related structure data
| Related structure data |  8gafMC  8g9sC  8g9tC  8g9uC  8gamC  8ganC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29896.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29896.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.4057 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29896_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29896_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of Cas3 with dsDNA bound type I-C Cascade at R-lo...
| Entire | Name: Ternary complex of Cas3 with dsDNA bound type I-C Cascade at R-loop formation |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of Cas3 with dsDNA bound type I-C Cascade at R-lo...
| Supramolecule | Name: Ternary complex of Cas3 with dsDNA bound type I-C Cascade at R-loop formation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Cas7
| Macromolecule | Name: Cas7 / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) |
| Molecular weight | Theoretical: 32.208111 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: TIEKRYDFVF LFDVQDGNPN GDPDAGNLPR IDPQTGEGLV TDVCLKRKVR NFIQMTQNDE HHDIFIREKG ILNNLIDEAH EQENVKGKE KGEKTEAARQ YMCSRYYDIR TFGAVMTTGK NAGQVRGPVQ LTFSRSIDPI MTLEHSITRM AVTNEKDASE T GDNRTMGR ...String: TIEKRYDFVF LFDVQDGNPN GDPDAGNLPR IDPQTGEGLV TDVCLKRKVR NFIQMTQNDE HHDIFIREKG ILNNLIDEAH EQENVKGKE KGEKTEAARQ YMCSRYYDIR TFGAVMTTGK NAGQVRGPVQ LTFSRSIDPI MTLEHSITRM AVTNEKDASE T GDNRTMGR KFTVPYGLYR CHGFISTHFA KQTGFSENDL ELFWQALVNM FDHDHSAARG QMNARGLYVF EHSNNLGDAP AD SLFKRIQ VVKKDGVEVV RSFDDYLVSV DDKNLEETKL LRKLGG UniProtKB:  UNIPROTKB: A0A378VEU0 UNIPROTKB: A0A378VEU0 |
-Macromolecule #2: Cas8
| Macromolecule | Name: Cas8 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) |
| Molecular weight | Theoretical: 64.715652 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MILHALTQYY QRKAESDGGI AQEGFENKEI PFIIVIDKQG NFIQLEDTRE LKVKKKVGRT FLVPKGLGRS GSKSYEVSNL LWDHYGYVL AYAGEKGQEQ ADKQHASFTA KVNELKQALP DDAGVTAVAA FLSSAEEKSK VMQAANWAEC AKVKGCNLSF R LVDEAVDL ...String: MILHALTQYY QRKAESDGGI AQEGFENKEI PFIIVIDKQG NFIQLEDTRE LKVKKKVGRT FLVPKGLGRS GSKSYEVSNL LWDHYGYVL AYAGEKGQEQ ADKQHASFTA KVNELKQALP DDAGVTAVAA FLSSAEEKSK VMQAANWAEC AKVKGCNLSF R LVDEAVDL VCQSKAVREY VSQANQTQSD NAQKGICLVT GKAAPIARLH NAVKGVNAKP APFASVNLSA FESYGKEQGF AF PIGEQAM FEYTTALNTL LAGENRFRIG DVTTVCWGAK RTPLEESLAS LINGGGKDKP DAHIDAVKAL YKSLYNGQYC KPD GEDKFY LLGLSPNSAR IVVRFWHETT VAALSESIAA WYDDLQMVRG ENSPYPEYMP LPRLLGNLVL DGKMENLPSD LIAQ ITDAA LNNRVLPVSL LQAALRRNKA EQKITYGRAS LLKAYINRAI RAGRLKNMKE LTMGLDRNRQ DIGYVLGRLF AVLEK IQAE ANPGLNATIA DRYFGSASST PIAVFGTLMR LLPHHLNKLE FEGRAVQLQW EIRQILEHCQ RFPNHLNLEQ QGLFAI GYY HETQFLFTKD ALKNLFNEA UniProtKB: Type I-C CRISPR-associated protein Cas8c/Csd1 |
-Macromolecule #3: Cas11
| Macromolecule | Name: Cas11 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) |
| Molecular weight | Theoretical: 14.245184 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GLDRNRQDIG YVLGRLFAVL EKIQAEANPG LNATIADRYF GSASSTPIAV FGTLMRLLPH HLNKLEFEGR AVQLQWEIRQ ILEHCQRFP NHLNLEQQGL FAIGYYHETQ FLFTKDALKN LFNEA UniProtKB: UNIPROTKB: A0A378VF47 |
-Macromolecule #5: Cas5
| Macromolecule | Name: Cas5 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) |
| Molecular weight | Theoretical: 23.870451 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: RFILEISGDL ACFTRSELKV ERVSYPVITP SAARNILMAI LWKPAIRWKV LKIEILKPIQ WTNIRRNEVG TKMSERSGSL YIEDNRQQR ASMLLKDVAY RIHADFDMTS EAGESDNYVK FAEMFKRRAK KGQYFHQPYL GCREFPCDFR LLEKAEDGLP L EDITQDFG ...String: RFILEISGDL ACFTRSELKV ERVSYPVITP SAARNILMAI LWKPAIRWKV LKIEILKPIQ WTNIRRNEVG TKMSERSGSL YIEDNRQQR ASMLLKDVAY RIHADFDMTS EAGESDNYVK FAEMFKRRAK KGQYFHQPYL GCREFPCDFR LLEKAEDGLP L EDITQDFG FMLYDMDFSK SDPRDSNNAE PMFYQCKAVN GVITVPP UniProtKB: pre-crRNA processing endonuclease |
-Macromolecule #4: crRNA (43-MER)
| Macromolecule | Name: crRNA (43-MER) / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Neisseria lactamica (bacteria) Neisseria lactamica (bacteria) |
| Molecular weight | Theoretical: 13.816202 KDa |
| Sequence | String: GUUGAAACAG GGUCAGCUUG CCGUAGGUGG CAUCGCCCUC GUC |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 150.0 mM / Component - Formula: NaCl Sodium chloride / Component - Name: sodium chloride Sodium chloride / Component - Name: sodium chloride / Details: 25mM Tris pH 7.5, 150mM NaCl / Details: 25mM Tris pH 7.5, 150mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 67000 / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 67000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 70.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1798 / Number real images: 1798 / Average exposure time: 2.5 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: NOT APPLICABLE |
| Final 3D classification | Number classes: 6 / Software - Name: cryoSPARC |
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.64 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 103405 |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8gaf: |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X

















