[English] 日本語
 Yorodumi
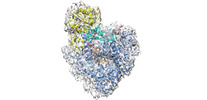
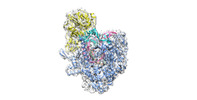
Yorodumi- PDB-6ymw: Cryo-EM structure of yeast mitochondrial RNA polymerase transcrip... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ymw | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TRANSCRIPTION / TRANSCRIPTION /  gene transcription / gene transcription /  polymerase / polymerase /  RDRP / MTF1 / RPO41 / RDRP / MTF1 / RPO41 /  POLRMT / mtRNAP / POLRMT / mtRNAP /  DNA / DNA /  transcription initiation / transcription initiation /  RNA polymerase / RNA polymerase /  mitochondria mitochondria | ||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial transcription factor activity /  transcription initiation at mitochondrial promoter / mitochondrial promoter sequence-specific DNA binding / rRNA (adenine-N6,N6-)-dimethyltransferase activity / mitochondrial transcription / transcription initiation at mitochondrial promoter / mitochondrial promoter sequence-specific DNA binding / rRNA (adenine-N6,N6-)-dimethyltransferase activity / mitochondrial transcription /  mitochondrial genome maintenance / mitochondrial genome maintenance /  DNA primase activity / DNA primase activity /  DNA replication, synthesis of primer ...Mitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial transcription factor activity / DNA replication, synthesis of primer ...Mitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial transcription factor activity /  transcription initiation at mitochondrial promoter / mitochondrial promoter sequence-specific DNA binding / rRNA (adenine-N6,N6-)-dimethyltransferase activity / mitochondrial transcription / transcription initiation at mitochondrial promoter / mitochondrial promoter sequence-specific DNA binding / rRNA (adenine-N6,N6-)-dimethyltransferase activity / mitochondrial transcription /  mitochondrial genome maintenance / mitochondrial genome maintenance /  DNA primase activity / DNA primase activity /  DNA replication, synthesis of primer / positive regulation of DNA-templated transcription, elongation / mitochondrial nucleoid / DNA replication, synthesis of primer / positive regulation of DNA-templated transcription, elongation / mitochondrial nucleoid /  Transferases; Transferring one-carbon groups; Methyltransferases / Transferases; Transferring one-carbon groups; Methyltransferases /  mitochondrial intermembrane space / DNA-directed 5'-3' RNA polymerase activity / mitochondrial intermembrane space / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / DNA-directed RNA polymerase /  mitochondrial matrix / mitochondrial matrix /  mitochondrion / mitochondrion /  DNA binding / DNA binding /  RNA binding / RNA binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)  Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast)synthetic construct (others) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.71 Å cryo EM / Resolution: 3.71 Å | ||||||
 Authors Authors | Das, K. / Patel, S.S. | ||||||
| Funding support |  Belgium, 1items Belgium, 1items
| ||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Cryo-EM Structures Reveal Transcription Initiation Steps by Yeast Mitochondrial RNA Polymerase. Authors: Brent De Wijngaert / Shemaila Sultana / Anupam Singh / Chhaya Dharia / Hans Vanbuel / Jiayu Shen / Daniel Vasilchuk / Sergio E Martinez / Eaazhisai Kandiah / Smita S Patel / Kalyan Das /    Abstract: Mitochondrial RNA polymerase (mtRNAP) is crucial in cellular energy production, yet understanding of mitochondrial DNA transcription initiation lags that of bacterial and nuclear DNA transcription. ...Mitochondrial RNA polymerase (mtRNAP) is crucial in cellular energy production, yet understanding of mitochondrial DNA transcription initiation lags that of bacterial and nuclear DNA transcription. We report structures of two transcription initiation intermediate states of yeast mtRNAP that explain promoter melting, template alignment, DNA scrunching, abortive synthesis, and transition into elongation. In the partially melted initiation complex (PmIC), transcription factor MTF1 makes base-specific interactions with flipped non-template (NT) nucleotides "AAGT" at -4 to -1 positions of the DNA promoter. In the initiation complex (IC), the template in the expanded 7-mer bubble positions the RNA and NTP analog UTPαS, while NT scrunches into an NT loop. The scrunched NT loop is stabilized by the centrally positioned MTF1 C-tail. The IC and PmIC states coexist in solution, revealing a dynamic equilibrium between two functional states. Frequent scrunching/unscruching transitions and the imminent steric clashes of the inflating NT loop and growing RNA:DNA with the C-tail explain abortive synthesis and transition into elongation. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ymw.cif.gz 6ymw.cif.gz | 283 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ymw.ent.gz pdb6ymw.ent.gz | 209.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ymw.json.gz 6ymw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ym/6ymw https://data.pdbj.org/pub/pdb/validation_reports/ym/6ymw ftp://data.pdbj.org/pub/pdb/validation_reports/ym/6ymw ftp://data.pdbj.org/pub/pdb/validation_reports/ym/6ymw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  10846MC  6ymvC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 2 molecules BA
| #1: Protein | Mass: 41151.203 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast)Strain: ATCC 204508 / S288c / Gene: MTF1, YMR228W, YM9959.10 / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: P14908,  Transferases; Transferring one-carbon groups; Methyltransferases Transferases; Transferring one-carbon groups; Methyltransferases |
|---|---|
| #2: Protein |  Polymerase PolymeraseMass: 143282.656 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast)Strain: ATCC 204508 / S288c / Gene: RPO41, YFL036W / Production host:   Escherichia coli (E. coli) / References: UniProt: P13433, Escherichia coli (E. coli) / References: UniProt: P13433,  DNA-directed RNA polymerase DNA-directed RNA polymerase |
-DNA chain , 2 types, 2 molecules NT
| #3: DNA chain | Mass: 10248.671 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: 15S mitochondria / Source: (synth.)   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) |
|---|---|
| #4: DNA chain | Mass: 10047.511 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: 15S mitochondria / Source: (synth.)   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) |
-RNA chain , 1 types, 1 molecules C
| #5: RNA chain | Mass: 805.413 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|
-Non-polymers , 2 types, 2 molecules 


| #6: Chemical | ChemComp-P5E / [[( |
|---|---|
| #7: Chemical | ChemComp-MG / |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.202 MDa / Experimental value: NO | ||||||||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||||||||
| Source (recombinant) |
| ||||||||||||||||||||||||||||||
| Buffer solution | pH: 7 Details: 50mM Bis-tris propane, 100mM NaCl, 5mM MgCl2, 1mM EDTA, 2mM DTT | ||||||||||||||||||||||||||||||
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||||||||
Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 95 % / Chamber temperature: 8 K / Details: 5 uL sample; back blotting for 12 -14 second |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 165000 X / Nominal defocus max: 22000 nm / Nominal defocus min: 7000 nm / Calibrated defocus min: 5500 nm / Calibrated defocus max: 26000 nm / Cs Bright-field microscopy / Nominal magnification: 165000 X / Nominal defocus max: 22000 nm / Nominal defocus min: 7000 nm / Calibrated defocus min: 5500 nm / Calibrated defocus max: 26000 nm / Cs : 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: COMA FREE : 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 6 sec. / Electron dose: 61 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 / Num. of real images: 3500 |
| EM imaging optics | Energyfilter slit width: 20 eV |
| Image scans | Movie frames/image: 50 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 3.71 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 62807 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL / Target criteria: Coorelation coefficu=ient / Details: Realspace refinement | ||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Source name: PDB / Type: experimental model
| ||||||||||||||||||||||||||||||||||||
| Refinement | Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 55.32 Å2 | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller

























 PDBj
PDBj









































