+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | State 2 Yeast V-ATPase without Oxr1p bound | |||||||||
 Map data Map data | Yeast V-ATPase in state 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Vacuolar ATPase / Vacuolar ATPase /  Proton pump / Oxr1p / Proton pump / Oxr1p /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
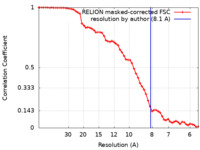
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 8.1 Å cryo EM / Resolution: 8.1 Å | |||||||||
 Authors Authors | Khan MM / Wilkens S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2024 Journal: EMBO Rep / Year: 2024Title: Molecular mechanism of Oxr1p mediated disassembly of yeast V-ATPase. Authors: Md Murad Khan / Stephan Wilkens /  Abstract: The eukaryotic vacuolar H-ATPase (V-ATPase) is regulated by reversible disassembly into autoinhibited V-ATPase and V proton channel subcomplexes. We recently reported that the TLDc protein Oxr1p ...The eukaryotic vacuolar H-ATPase (V-ATPase) is regulated by reversible disassembly into autoinhibited V-ATPase and V proton channel subcomplexes. We recently reported that the TLDc protein Oxr1p induces V-ATPase disassembly in vitro. Whether and how Oxr1p is involved in enzyme disassembly in vivo, however, is not known. Here, using yeast genetics and fluorescence microscopy, we show that Oxr1p is essential for efficient V-ATPase disassembly in the cell. Supporting biochemical and biophysical in vitro experiments show that whereas Oxr1p-driven holoenzyme disassembly can occur in the absence of nucleotides, the presence of ATP greatly accelerates the process. ATP hydrolysis is needed, however, for subsequent release of Oxr1p so that the free V can adopt the autoinhibited conformation. Overall, our study unravels the molecular mechanism of Oxr1p-induced disassembly that occurs in vivo as part of the canonical V-ATPase regulation by reversible disassembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42881.map.gz emd_42881.map.gz | 11.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42881-v30.xml emd-42881-v30.xml emd-42881.xml emd-42881.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42881_fsc.xml emd_42881_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42881.png emd_42881.png | 73.1 KB | ||
| Masks |  emd_42881_msk_1.map emd_42881_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42881.cif.gz emd-42881.cif.gz | 3.9 KB | ||
| Others |  emd_42881_half_map_1.map.gz emd_42881_half_map_1.map.gz emd_42881_half_map_2.map.gz emd_42881_half_map_2.map.gz | 11.9 MB 11.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42881 http://ftp.pdbj.org/pub/emdb/structures/EMD-42881 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42881 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42881 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42881.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42881.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast V-ATPase in state 2 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 2.85 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42881_msk_1.map emd_42881_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_42881_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_42881_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast Vacuolar ATPase vitrified in presence of Oxr1p
| Entire | Name: Yeast Vacuolar ATPase vitrified in presence of Oxr1p |
|---|---|
| Components |
|
-Supramolecule #1: Yeast Vacuolar ATPase vitrified in presence of Oxr1p
| Supramolecule | Name: Yeast Vacuolar ATPase vitrified in presence of Oxr1p / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / Organelle: Vacuole Saccharomyces cerevisiae (brewer's yeast) / Organelle: Vacuole |
| Molecular weight | Theoretical: 1 MDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: TBS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 279 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X