[English] 日本語
 Yorodumi
Yorodumi- EMDB-40237: Structure of wild-type MCM8/9 heterohexamer complex with ATP-gamma-S -
+ Open data
Open data
- Basic information
Basic information
| Entry | 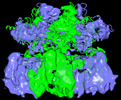 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of wild-type MCM8/9 heterohexamer complex with ATP-gamma-S | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  DNA Replication / DNA Replication /  DNA repair / DNA repair /  DNA BINDING PROTEIN DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMutLbeta complex binding / MutSbeta complex binding / recombinational interstrand cross-link repair / MCM8-MCM9 complex / male gamete generation / mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication / CDC6 association with the ORC:origin complex / MutSalpha complex binding / E2F-enabled inhibition of pre-replication complex formation / female gamete generation ...MutLbeta complex binding / MutSbeta complex binding / recombinational interstrand cross-link repair / MCM8-MCM9 complex / male gamete generation / mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication / CDC6 association with the ORC:origin complex / MutSalpha complex binding / E2F-enabled inhibition of pre-replication complex formation / female gamete generation / Unwinding of DNA / MCM complex / DNA duplex unwinding / Activation of the pre-replicative complex / Activation of ATR in response to replication stress / protein localization to chromatin /  DNA helicase activity / DNA helicase activity /  helicase activity / double-strand break repair via homologous recombination / Orc1 removal from chromatin / helicase activity / double-strand break repair via homologous recombination / Orc1 removal from chromatin /  single-stranded DNA binding / single-stranded DNA binding /  chromosome / chromosome /  DNA helicase / protein stabilization / DNA helicase / protein stabilization /  cell cycle / DNA damage response / cell cycle / DNA damage response /  chromatin binding / protein-containing complex binding / chromatin binding / protein-containing complex binding /  enzyme binding / enzyme binding /  ATP hydrolysis activity / ATP hydrolysis activity /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  nucleus nucleusSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.5 Å cryo EM / Resolution: 6.5 Å | |||||||||
 Authors Authors | Li C / Gao Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Activity, substrate preference and structure of the HsMCM8/9 helicase. Authors: David R McKinzey / Chuxuan Li / Yang Gao / Michael A Trakselis /  Abstract: The minichromosomal maintenance proteins, MCM8 and MCM9, are more recent evolutionary additions to the MCM family, only cooccurring in selected higher eukaryotes. Mutations in these genes are ...The minichromosomal maintenance proteins, MCM8 and MCM9, are more recent evolutionary additions to the MCM family, only cooccurring in selected higher eukaryotes. Mutations in these genes are directly linked to ovarian insufficiency, infertility, and several cancers. MCM8/9 appears to have ancillary roles in fork progression and recombination of broken replication forks. However, the biochemical activity, specificities and structures have not been adequately illustrated, making mechanistic determination difficult. Here, we show that human MCM8/9 (HsMCM8/9) is an ATP dependent DNA helicase that unwinds fork DNA substrates with a 3'-5' polarity. High affinity ssDNA binding occurs in the presence of nucleoside triphosphates, while ATP hydrolysis weakens the interaction with DNA. The cryo-EM structure of the HsMCM8/9 heterohexamer was solved at 4.3 Å revealing a trimer of heterodimer configuration with two types of interfacial AAA+ nucleotide binding sites that become more organized upon binding ADP. Local refinements of the N or C-terminal domains (NTD or CTD) improved the resolution to 3.9 or 4.1 Å, respectively, and shows a large displacement in the CTD. Changes in AAA+ CTD upon nucleotide binding and a large swing between the NTD and CTD likely implies that MCM8/9 utilizes a sequential subunit translocation mechanism for DNA unwinding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
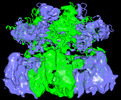
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40237.map.gz emd_40237.map.gz | 117.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40237-v30.xml emd-40237-v30.xml emd-40237.xml emd-40237.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40237.png emd_40237.png | 179.2 KB | ||
| Others |  emd_40237_half_map_1.map.gz emd_40237_half_map_1.map.gz emd_40237_half_map_2.map.gz emd_40237_half_map_2.map.gz | 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40237 http://ftp.pdbj.org/pub/emdb/structures/EMD-40237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40237 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40237 | HTTPS FTP |
-Related structure data
| Related structure data |  8s91C  8s92C  8s94C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40237.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40237.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_40237_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40237_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of wild-type MCM8/9 heterohexamer complex with ATP-gamma-S
| Entire | Name: Structure of wild-type MCM8/9 heterohexamer complex with ATP-gamma-S |
|---|---|
| Components |
|
-Supramolecule #1: Structure of wild-type MCM8/9 heterohexamer complex with ATP-gamma-S
| Supramolecule | Name: Structure of wild-type MCM8/9 heterohexamer complex with ATP-gamma-S type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: MCM8_HUMAN
| Macromolecule | Name: MCM8_HUMAN / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MNGEYRGRGF GRGRFQSWKR GRGGGNFSGK WREREHRPDL SKTTGKRTSE QTPQFLLSTK TPQSMQSTLD RFIPYKGWKL YFSEVYSDSS PLIEKIQAFE KFFTRHIDLY DKDEIERKGS ILVDFKELTE GGEVTNLIPD IATELRDAPE KTLACMGLAI HQVLTKDLER ...String: MNGEYRGRGF GRGRFQSWKR GRGGGNFSGK WREREHRPDL SKTTGKRTSE QTPQFLLSTK TPQSMQSTLD RFIPYKGWKL YFSEVYSDSS PLIEKIQAFE KFFTRHIDLY DKDEIERKGS ILVDFKELTE GGEVTNLIPD IATELRDAPE KTLACMGLAI HQVLTKDLER HAAELQAQEG LSNDGETMVN VPHIHARVYN YEPLTQLKNV RANYYGKYIA LRGTVVRVSN IKPLCTKMAF LCAACGEIQS FPLPDGKYSL PTKCPVPVCR GRSFTALRSS PLTVTMDWQS IKIQELMSDD QREAGRIPRT IECELVHDLV DSCVPGDTVT ITGIVKVSNA EEGSRNKNDK CMFLLYIEAN SISNSKGQKT KSSEDGCKHG MLMEFSLKDL YAIQEIQAEE NLFKLIVNSL CPVIFGHELV KAGLALALFG GSQKYADDKN RIPIRGDPHI LVVGDPGLGK SQMLQAACNV APRGVYVCGN TTTTSGLTVT LSKDSSSGDF ALEAGALVLG DQGICGIDEF DKMGNQHQAL LEAMEQQSIS LAKAGVVCSL PARTSIIAAA NPVGGHYNKA KTVSENLKMG SALLSRFDLV FILLDTPNEH HDHLLSEHVI AIRAGKQRTI SSATVARMNS QDSNTSVLEV VSEKPLSERL KVVPGETIDP IPHQLLRKYI GYARQYVYPR LSTEAARVLQ DFYLELRKQS QRLNSSPITT RQLESLIRLT EARARLELRE EATKEDAEDI VEIMKYSMLG TYSDEFGNLD FERSQHGSGM SNRSTAKRFI SALNNVAERT YNNIFQFHQL RQIAKELNIQ VADFENFIGS LNDQGYLLKK GPKVYQLQTM UniProtKB: DNA helicase MCM8 |
-Macromolecule #2: MCM9_HUMAN
| Macromolecule | Name: MCM9_HUMAN / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MNSDQVTLVG QVFESYVSEY HKNDILLILK ERDEDAHYPV VVNAMTLFET NMEIGEYFNM FPSEVLTIFD SALRRSALTI LQSLSQPEAV SMKQNLHARI SGLPVCPELV REHIPKTKDV GHFLSVTGTV IRTSLVKVLE FERDYMCNKC KHVFVIKADF EQYYTFCRPS ...String: MNSDQVTLVG QVFESYVSEY HKNDILLILK ERDEDAHYPV VVNAMTLFET NMEIGEYFNM FPSEVLTIFD SALRRSALTI LQSLSQPEAV SMKQNLHARI SGLPVCPELV REHIPKTKDV GHFLSVTGTV IRTSLVKVLE FERDYMCNKC KHVFVIKADF EQYYTFCRPS SCPSLESCDS SKFTCLSGLS SSPTRCRDYQ EIKIQEQVQR LSVGSIPRSM KVILEDDLVD SCKSGDDLTI YGIVMQRWKP FQQDVRCEVE IVLKANYIQV NNEQSSGIIM DEEVQKEFED FWEYYKSDPF AGRNVILASL CPQVFGMYLV KLAVAMVLAG GIQRTDATGT RVRGESHLLL VGDPGTGKSQ FLKYAAKITP RSVLTTGIGS TSAGLTVTAV KDSGEWNLEA GALVLADAGL CCIDEFNSLK EHDRTSIHEA MEQQTISVAK AGLVCKLNTR TTILAATNPK GQYDPQESVS VNIALGSPLL SRFDLILVLL DTKNEDWDRI ISSFILENKG YPSKSEKLWS MEKMKTYFCL IRNLQPTLSD VGNQVLLRYY QMQRQSDCRN AARTTIRLLE SLIRLAEAHA RLMFRDTVTL EDAITVVSVM ESSMQGGALL GGVNALHTSF PENPGEQYQR QCELILEKLE LQSLLSEELR RLERLQNQSV HQSQPRVLEV ETTPGSLRNG PGEESNFRTS SQQEINYSTH IFSPGGSPEG SPVLDPPPHL EPNRSTSRKH SAQHKNNRDD SLDWFDFMAT HQSEPKNTVV VSPHPKTSGE NMASKISNST SQGKEKSEPG QRSKVDIGLL PSPGETGVPW RADNVESNKK KRLALDSEAA VSADKPDSVL THHVPRNLQK LCKERAQKLC RNSTRVPAQC TVPSHPQSTP VHSPDRMLDS PKRKRPKSLA QVEEPAIENV KPPGSPVAKL AKFTFKQKSK LIHSFEDHSH VSPGATKIAV HSPKISQRRT RRDAALPVKR PGKLTSTPGN QISSQPQGET KEVSQQPPEK HGPREKVMCA PEKRIIQPEL ELGNETGCAH LTCEGDKKEE VSGSNKSGKV HACTLARLAN FCFTPPSESK SKSPPPERKN RGERGPSSPP TTTAPMRVSK RKSFQLRGST EKLIVSKESL FTLPELGDEA FDCDWDEEMR KKS UniProtKB: DNA helicase MCM9 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.7000000000000001 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 49.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 57384 |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X