[English] 日本語
 Yorodumi
Yorodumi- EMDB-33931: Structure of photosynthetic LH1-RC super-complex of Rhodobacter c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of photosynthetic LH1-RC super-complex of Rhodobacter capsulatus | |||||||||||||||||||||
 Map data Map data | negative B-factor applied map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex /  bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II /  photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||||||||||||||
| Biological species |   Rhodobacter capsulatus (bacteria) Rhodobacter capsulatus (bacteria) | |||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.6 Å cryo EM / Resolution: 2.6 Å | |||||||||||||||||||||
 Authors Authors | Tani K / Kanno R / Ji X-C / Satoh I / Kobayashi Y / Nagashima KVP / Hall M / Yu L-J / Kimura Y / Mizoguchi A ...Tani K / Kanno R / Ji X-C / Satoh I / Kobayashi Y / Nagashima KVP / Hall M / Yu L-J / Kimura Y / Mizoguchi A / Humbel BM / Madigan MT / Wang-Otomo Z-Y | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Rhodobacter capsulatus forms a compact crescent-shaped LH1-RC photocomplex. Authors: Kazutoshi Tani / Ryo Kanno / Xuan-Cheng Ji / Itsusei Satoh / Yuki Kobayashi / Malgorzata Hall / Long-Jiang Yu / Yukihiro Kimura / Akira Mizoguchi / Bruno M Humbel / Michael T Madigan / Zheng-Yu Wang-Otomo /    Abstract: Rhodobacter (Rba.) capsulatus has been a favored model for studies of all aspects of bacterial photosynthesis. This purple phototroph contains PufX, a polypeptide crucial for dimerization of the ...Rhodobacter (Rba.) capsulatus has been a favored model for studies of all aspects of bacterial photosynthesis. This purple phototroph contains PufX, a polypeptide crucial for dimerization of the light-harvesting 1-reaction center (LH1-RC) complex, but lacks protein-U, a U-shaped polypeptide in the LH1-RC of its close relative Rba. sphaeroides. Here we present a cryo-EM structure of the Rba. capsulatus LH1-RC purified by DEAE chromatography. The crescent-shaped LH1-RC exhibits a compact structure containing only 10 LH1 αβ-subunits. Four αβ-subunits corresponding to those adjacent to protein-U in Rba. sphaeroides were absent. PufX in Rba. capsulatus exhibits a unique conformation in its N-terminus that self-associates with amino acids in its own transmembrane domain and interacts with nearby polypeptides, preventing it from interacting with proteins in other complexes and forming dimeric structures. These features are discussed in relation to the minimal requirements for the formation of LH1-RC monomers and dimers, the spectroscopic behavior of both the LH1 and RC, and the bioenergetics of energy transfer from LH1 to the RC. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33931.map.gz emd_33931.map.gz | 228.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33931-v30.xml emd-33931-v30.xml emd-33931.xml emd-33931.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33931_fsc.xml emd_33931_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33931.png emd_33931.png | 149.3 KB | ||
| Others |  emd_33931_half_map_1.map.gz emd_33931_half_map_1.map.gz emd_33931_half_map_2.map.gz emd_33931_half_map_2.map.gz | 194.6 MB 194.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33931 http://ftp.pdbj.org/pub/emdb/structures/EMD-33931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33931 | HTTPS FTP |
-Related structure data
| Related structure data |  7ymlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33931.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33931.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | negative B-factor applied map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half odd map
| File | emd_33931_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half odd map | ||||||||||||
| Projections & Slices |
| ||||||||||||
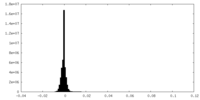
| Density Histograms |
-Half map: Half even map
| File | emd_33931_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half even map | ||||||||||||
| Projections & Slices |
| ||||||||||||
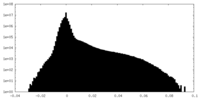
| Density Histograms |
- Sample components
Sample components
+Entire : Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Supramolecule #1: Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Macromolecule #1: Reaction center protein L chain
+Macromolecule #2: Reaction center protein M chain
+Macromolecule #3: Photosynthetic reaction center H subunit
+Macromolecule #4: Light-harvesting protein B-870 alpha chain
+Macromolecule #5: Light-harvesting protein B-870 beta chain
+Macromolecule #6: Photosynthetic reaction center PufX protein
+Macromolecule #7: BACTERIOCHLOROPHYLL A
+Macromolecule #8: BACTERIOPHEOPHYTIN A
+Macromolecule #9: UBIQUINONE-10
+Macromolecule #10: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #11: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
+Macromolecule #12: LAURYL DIMETHYLAMINE-N-OXIDE
+Macromolecule #13: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #14: FE (III) ION
+Macromolecule #15: SPHEROIDENE
+Macromolecule #16: MYRISTIC ACID
+Macromolecule #17: water
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 31.8 sec. / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X