+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
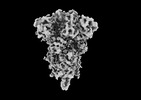
| Title | Subtomogram average of SARS-CoV-2 Delta variant postfusion S. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 | |||||||||
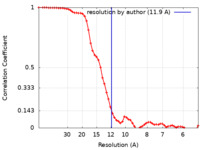
| Method | subtomogram averaging /  cryo EM / Resolution: 11.9 Å cryo EM / Resolution: 11.9 Å | |||||||||
 Authors Authors | Song Y / Zhang Z / Peng C / Li S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: In situ architecture and membrane fusion of SARS-CoV-2 Delta variant. Authors: Yutong Song / Hangping Yao / Nanping Wu / Jialu Xu / Zheyuan Zhang / Cheng Peng / Shibo Li / Weizheng Kong / Yong Chen / Miaojin Zhu / Jiaqi Wang / Danrong Shi / Chongchong Zhao / Xiangyun ...Authors: Yutong Song / Hangping Yao / Nanping Wu / Jialu Xu / Zheyuan Zhang / Cheng Peng / Shibo Li / Weizheng Kong / Yong Chen / Miaojin Zhu / Jiaqi Wang / Danrong Shi / Chongchong Zhao / Xiangyun Lu / Martín Echavarría Galindo / Sai Li /  Abstract: Among the current five Variants of Concern, infections caused by SARS-CoV-2 B.1.617.2 (Delta) variant are often associated with the greatest severity. Despite recent advances on the molecular basis ...Among the current five Variants of Concern, infections caused by SARS-CoV-2 B.1.617.2 (Delta) variant are often associated with the greatest severity. Despite recent advances on the molecular basis of elevated pathogenicity using recombinant proteins, the architecture of intact Delta virions remains veiled. Moreover, pieces of molecular evidence for the detailed mechanism of S-mediated membrane fusion are missing. Here, we showed the pleomorphic nature of Delta virions from electron beam inactivated samples and reported the in situ structure and distribution of S on the authentic Delta variant. We also captured the virus-virus fusion events, which provided pieces of structural evidence for Delta's attenuated dependency on cellular factors for fusion activation, and proposed a model of S-mediated membrane fusion. Besides, site-specific glycan analysis revealed increased oligomannose-type glycosylation of native Delta S than that of the WT S. Together, these results disclose distinctive factors of Delta being the most virulent SARS-CoV-2 variant. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33208.map.gz emd_33208.map.gz | 14.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33208-v30.xml emd-33208-v30.xml emd-33208.xml emd-33208.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33208_fsc.xml emd_33208_fsc.xml | 6.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33208.png emd_33208.png | 51.1 KB | ||
| Others |  emd_33208_half_map_1.map.gz emd_33208_half_map_1.map.gz emd_33208_half_map_2.map.gz emd_33208_half_map_2.map.gz | 14.5 MB 14.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33208 http://ftp.pdbj.org/pub/emdb/structures/EMD-33208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33208 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33208 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33208.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33208.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 2.72 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33208_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33208_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Severe acute respiratory syndrome coronavirus 2
| Entire | Name:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
|---|---|
| Components |
|
-Supramolecule #1: Severe acute respiratory syndrome coronavirus 2
| Supramolecule | Name: Severe acute respiratory syndrome coronavirus 2 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 2697049 Sci species name: Severe acute respiratory syndrome coronavirus 2 Sci species strain: B.1.617.2 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 64000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 64000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average exposure time: 0.213 sec. / Average electron dose: 3.2 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X