[English] 日本語
 Yorodumi
Yorodumi- EMDB-19255: Human Replication protein A (RPA; trimeric core) - ssDNA complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Replication protein A (RPA; trimeric core) - ssDNA complex DNA replication DNA replication | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | ssDNA binding protein /  DNA damage repair / Single-strand annealing / DNA damage repair / Single-strand annealing /  DNA BINDING PROTEIN DNA BINDING PROTEIN | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to chromosome / DNA replication factor A complex / chromatin-protein adaptor activity / protein localization to site of double-strand break /  regulation of DNA damage checkpoint / Removal of the Flap Intermediate / single-stranded telomeric DNA binding / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Removal of the Flap Intermediate from the C-strand ...protein localization to chromosome / DNA replication factor A complex / chromatin-protein adaptor activity / protein localization to site of double-strand break / regulation of DNA damage checkpoint / Removal of the Flap Intermediate / single-stranded telomeric DNA binding / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Removal of the Flap Intermediate from the C-strand ...protein localization to chromosome / DNA replication factor A complex / chromatin-protein adaptor activity / protein localization to site of double-strand break /  regulation of DNA damage checkpoint / Removal of the Flap Intermediate / single-stranded telomeric DNA binding / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Removal of the Flap Intermediate from the C-strand / G-rich strand telomeric DNA binding / HDR through Single Strand Annealing (SSA) / Impaired BRCA2 binding to RAD51 / regulation of double-strand break repair via homologous recombination / telomeric DNA binding / site of DNA damage / Presynaptic phase of homologous DNA pairing and strand exchange / telomere maintenance via telomerase / PCNA-Dependent Long Patch Base Excision Repair / Activation of the pre-replicative complex / HSF1 activation / Regulation of HSF1-mediated heat shock response / regulation of DNA damage checkpoint / Removal of the Flap Intermediate / single-stranded telomeric DNA binding / Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Removal of the Flap Intermediate from the C-strand / G-rich strand telomeric DNA binding / HDR through Single Strand Annealing (SSA) / Impaired BRCA2 binding to RAD51 / regulation of double-strand break repair via homologous recombination / telomeric DNA binding / site of DNA damage / Presynaptic phase of homologous DNA pairing and strand exchange / telomere maintenance via telomerase / PCNA-Dependent Long Patch Base Excision Repair / Activation of the pre-replicative complex / HSF1 activation / Regulation of HSF1-mediated heat shock response /  mismatch repair / Activation of ATR in response to replication stress / SUMOylation of DNA damage response and repair proteins / mitotic G1 DNA damage checkpoint signaling / regulation of mitotic cell cycle / mismatch repair / Activation of ATR in response to replication stress / SUMOylation of DNA damage response and repair proteins / mitotic G1 DNA damage checkpoint signaling / regulation of mitotic cell cycle /  telomere maintenance / Translesion synthesis by REV1 / Translesion synthesis by POLK / Gap-filling DNA repair synthesis and ligation in GG-NER / Translesion synthesis by POLI / meiotic cell cycle / nucleotide-excision repair / Recognition of DNA damage by PCNA-containing replication complex / Fanconi Anemia Pathway / Termination of translesion DNA synthesis / double-strand break repair via homologous recombination / Translesion Synthesis by POLH / telomere maintenance / Translesion synthesis by REV1 / Translesion synthesis by POLK / Gap-filling DNA repair synthesis and ligation in GG-NER / Translesion synthesis by POLI / meiotic cell cycle / nucleotide-excision repair / Recognition of DNA damage by PCNA-containing replication complex / Fanconi Anemia Pathway / Termination of translesion DNA synthesis / double-strand break repair via homologous recombination / Translesion Synthesis by POLH /  base-excision repair / HDR through Homologous Recombination (HRR) / G2/M DNA damage checkpoint / Dual Incision in GG-NER / PML body / DNA-templated DNA replication / base-excision repair / HDR through Homologous Recombination (HRR) / G2/M DNA damage checkpoint / Dual Incision in GG-NER / PML body / DNA-templated DNA replication /  Meiotic recombination / Formation of Incision Complex in GG-NER / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / Meiotic recombination / Formation of Incision Complex in GG-NER / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER /  single-stranded DNA binding / site of double-strand break / regulation of cell population proliferation / Processing of DNA double-strand break ends / single-stranded DNA binding / site of double-strand break / regulation of cell population proliferation / Processing of DNA double-strand break ends /  protein phosphatase binding / DNA recombination / Regulation of TP53 Activity through Phosphorylation / protein phosphatase binding / DNA recombination / Regulation of TP53 Activity through Phosphorylation /  DNA replication / DNA replication /  chromosome, telomeric region / damaged DNA binding / chromosome, telomeric region / damaged DNA binding /  nuclear body / nuclear body /  DNA repair / DNA damage response / DNA repair / DNA damage response /  ubiquitin protein ligase binding / ubiquitin protein ligase binding /  chromatin / chromatin /  enzyme binding / enzyme binding /  nucleoplasm / nucleoplasm /  metal ion binding / metal ion binding /  nucleus nucleusSimilarity search - Function | |||||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
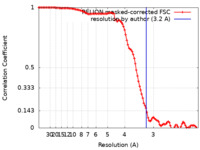
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||||||||
 Authors Authors | Liang CC / West SC | |||||||||||||||||||||||||||
| Funding support |  United Kingdom, European Union, United Kingdom, European Union,  Switzerland, 8 items Switzerland, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Mechanism of single-stranded DNA annealing by RAD52-RPA complex. Authors: Chih-Chao Liang / Luke A Greenhough / Laura Masino / Sarah Maslen / Ilirjana Bajrami / Marcel Tuppi / Mark Skehel / Ian A Taylor / Stephen C West /  Abstract: RAD52 is important for the repair of DNA double-stranded breaks, mitotic DNA synthesis and alternative telomere length maintenance. Central to these functions, RAD52 promotes the annealing of ...RAD52 is important for the repair of DNA double-stranded breaks, mitotic DNA synthesis and alternative telomere length maintenance. Central to these functions, RAD52 promotes the annealing of complementary single-stranded DNA (ssDNA) and provides an alternative to BRCA2/RAD51-dependent homologous recombination repair. Inactivation of RAD52 in homologous-recombination-deficient BRCA1- or BRCA2-defective cells is synthetically lethal, and aberrant expression of RAD52 is associated with poor cancer prognosis. As a consequence, RAD52 is an attractive therapeutic target against homologous-recombination-deficient breast, ovarian and prostate cancers. Here we describe the structure of RAD52 and define the mechanism of annealing. As reported previously, RAD52 forms undecameric (11-subunit) ring structures, but these rings do not represent the active form of the enzyme. Instead, cryo-electron microscopy and biochemical analyses revealed that ssDNA annealing is driven by RAD52 open rings in association with replication protein-A (RPA). Atomic models of the RAD52-ssDNA complex show that ssDNA sits in a positively charged channel around the ring. Annealing is driven by the RAD52 N-terminal domains, whereas the C-terminal regions modulate the open-ring conformation and RPA interaction. RPA associates with RAD52 at the site of ring opening with critical interactions occurring between the RPA-interacting domain of RAD52 and the winged helix domain of RPA2. Our studies provide structural snapshots throughout the annealing process and define the molecular mechanism of ssDNA annealing by the RAD52-RPA complex. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19255.map.gz emd_19255.map.gz | 6.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19255-v30.xml emd-19255-v30.xml emd-19255.xml emd-19255.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19255_fsc.xml emd_19255_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_19255.png emd_19255.png | 55.2 KB | ||
| Filedesc metadata |  emd-19255.cif.gz emd-19255.cif.gz | 6.8 KB | ||
| Others |  emd_19255_half_map_1.map.gz emd_19255_half_map_1.map.gz emd_19255_half_map_2.map.gz emd_19255_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19255 http://ftp.pdbj.org/pub/emdb/structures/EMD-19255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19255 | HTTPS FTP |
-Related structure data
| Related structure data |  8rk2MC  8rilC  8rj3C  8rjwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19255.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19255.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19255_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19255_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Open ring conformation of human RAD52 in complex with ssDNA
| Entire | Name: Open ring conformation of human RAD52 in complex with ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: Open ring conformation of human RAD52 in complex with ssDNA
| Supramolecule | Name: Open ring conformation of human RAD52 in complex with ssDNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinant RAD52 purified from E. coli, and the open ring conformation was separated by cation exchange. The RAD52-ssDNA complex was reconstituted in vitro. |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Replication protein A 70 kDa DNA-binding subunit, N-terminally pr...
| Macromolecule | Name: Replication protein A 70 kDa DNA-binding subunit, N-terminally processed type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.212977 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MVGQLSEGAI AAIMQKGDTN IKPILQVINI RPITTGNSPP RYRLLMSDGL NTLSSFMLAT QLNPLVEEEQ LSSNCVCQIH RFIVNTLKD GRRVVILMEL EVLKSAEAVG VKIGNPVPYN EGLGQPQVAP PAPAASPAAS SRPQPQNGSS GMGSTVSKAY G ASKTFGKA ...String: MVGQLSEGAI AAIMQKGDTN IKPILQVINI RPITTGNSPP RYRLLMSDGL NTLSSFMLAT QLNPLVEEEQ LSSNCVCQIH RFIVNTLKD GRRVVILMEL EVLKSAEAVG VKIGNPVPYN EGLGQPQVAP PAPAASPAAS SRPQPQNGSS GMGSTVSKAY G ASKTFGKA AGPSLSHTSG GTQSKVVPIA SLTPYQSKWT ICARVTNKSQ IRTWSNSRGE GKLFSLELVD ESGEIRATAF NE QVDKFFP LIEVNKVYYF SKGTLKIANK QFTAVKNDYE MTFNNETSVM PCEDDHHLPT VQFDFTGIDD LENKSKDSLV DII GICKSY EDATKITVRS NNREVAKRNI YLMDTSGKVV TATLWGEDAD KFDGSRQPVL AIKGARVSDF GGRSLSVLSS STII ANPDI PEAYKLRGWF DAEGQALDGV SISDLKSGGV GGSNTNWKTL YEVKSENLGQ GDKPDYFSSV ATVVYLRKEN CMYQA CPTQ DCNKKVIDQQ NGLYRCEKCD TEFPNFKYRM ILSVNIADFQ ENQWVTCFQE SAEAILGQNA AYLGELKDKN EQAFEE VFQ NANFRSFIFR VRVKVETYND ESRIKATVMD VKPVDYREYG RRLVMSIRRS ALM UniProtKB:  Replication protein A 70 kDa DNA-binding subunit Replication protein A 70 kDa DNA-binding subunit |
-Macromolecule #2: Replication protein A 32 kDa subunit
| Macromolecule | Name: Replication protein A 32 kDa subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.276795 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MWNSGFESYG SSSYGGAGGY TQSPGGFGSP APSQAEKKSR ARAQHIVPCT ISQLLSATLV DEVFRIGNVE ISQVTIVGII RHAEKAPTN IVYKIDDMTA APMDVRQWVD TDDTSSENTV VPPETYVKVA GHLRSFQNKK SLVAFKIMPL EDMNEFTTHI L EVINAHMV ...String: MWNSGFESYG SSSYGGAGGY TQSPGGFGSP APSQAEKKSR ARAQHIVPCT ISQLLSATLV DEVFRIGNVE ISQVTIVGII RHAEKAPTN IVYKIDDMTA APMDVRQWVD TDDTSSENTV VPPETYVKVA GHLRSFQNKK SLVAFKIMPL EDMNEFTTHI L EVINAHMV LSKANSQPSA GRAPISNPGM SEAGNFGGNS FMPANGLTVA QNQVLNLIKA CPRPEGLNFQ DLKNQLKHMS VS SIKQAVD FLSNEGHIYS TVDDDHFKST DAE UniProtKB:  Replication protein A 32 kDa subunit Replication protein A 32 kDa subunit |
-Macromolecule #3: Replication protein A 14 kDa subunit
| Macromolecule | Name: Replication protein A 14 kDa subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.583714 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MVDMMDLPRS RINAGMLAQF IDKPVCFVGR LEKIHPTGKM FILSDGEGKN GTIELMEPLD EEISGIVEVV GRVTAKATIL CTSYVQFKE DSHPFDLGLY NEAVKIIHDF PQFYPLGIVQ HD UniProtKB:  Replication protein A 14 kDa subunit Replication protein A 14 kDa subunit |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 46.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8rk2: |
 Movie
Movie Controller
Controller



















 Z
Z Y
Y X
X