[English] 日本語
 Yorodumi
Yorodumi- EMDB-19008: Cryo-EM structure of the human mPSF with PAPOA C-terminus peptide... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human mPSF with PAPOA C-terminus peptide (PAPOAc) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | 3-end processing /  polyadenylation / PAPOA / polyadenylation / PAPOA /  pre-mRNA / cleavage and polyadenylation / pre-mRNA / cleavage and polyadenylation /  RNA BINDING PROTEIN RNA BINDING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationco-transcriptional RNA 3'-end processing, cleavage and polyadenylation pathway / Inhibition of Host mRNA Processing and RNA Silencing / RNA 3'-end processing / Processing of Intronless Pre-mRNAs / mRNA cleavage and polyadenylation specificity factor complex / mRNA 3'-UTR AU-rich region binding / collagen trimer / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript /  postreplication repair ...co-transcriptional RNA 3'-end processing, cleavage and polyadenylation pathway / Inhibition of Host mRNA Processing and RNA Silencing / RNA 3'-end processing / Processing of Intronless Pre-mRNAs / mRNA cleavage and polyadenylation specificity factor complex / mRNA 3'-UTR AU-rich region binding / collagen trimer / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / postreplication repair ...co-transcriptional RNA 3'-end processing, cleavage and polyadenylation pathway / Inhibition of Host mRNA Processing and RNA Silencing / RNA 3'-end processing / Processing of Intronless Pre-mRNAs / mRNA cleavage and polyadenylation specificity factor complex / mRNA 3'-UTR AU-rich region binding / collagen trimer / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript /  postreplication repair / tRNA processing in the nucleus / RNA Polymerase II Transcription Termination / : / Processing of Capped Intron-Containing Pre-mRNA / postreplication repair / tRNA processing in the nucleus / RNA Polymerase II Transcription Termination / : / Processing of Capped Intron-Containing Pre-mRNA /  nucleotidyltransferase activity / nucleotidyltransferase activity /  fibrillar center / fibrillar center /  mRNA processing / sequence-specific double-stranded DNA binding / mRNA processing / sequence-specific double-stranded DNA binding /  spermatogenesis / intracellular membrane-bounded organelle / spermatogenesis / intracellular membrane-bounded organelle /  enzyme binding / enzyme binding /  RNA binding / zinc ion binding / RNA binding / zinc ion binding /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  nucleus nucleusSimilarity search - Function | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.79 Å cryo EM / Resolution: 2.79 Å | |||||||||||||||
 Authors Authors | Todesca S / Sandmeir F / Keidel A / Conti E | |||||||||||||||
| Funding support |  Germany, European Union, Germany, European Union,  Denmark, 4 items Denmark, 4 items
| |||||||||||||||
 Citation Citation |  Journal: RNA / Year: 2024 Journal: RNA / Year: 2024Title: Molecular basis of human polyA polymerase recruitment by mPSF. Authors: Sofia Todesca / Felix Sandmeir / Achim Keidel / Elena Conti Abstract: 3' end processing of most eukaryotic pre-mRNAs is a crucial co-transcriptional process that generally involves the cleavage and polyadenylation of the precursor transcripts. Within the human 3' end ...3' end processing of most eukaryotic pre-mRNAs is a crucial co-transcriptional process that generally involves the cleavage and polyadenylation of the precursor transcripts. Within the human 3' end processing machinery, the 4-subunit mammalian polyadenylation specificity factor (mPSF) recognizes the polyadenylation signal (PAS) in the pre-mRNA and recruits the polyA polymerase α (PAPOA) to it. To shed light on the molecular mechanisms of PAPOA recruitment to mPSF, we used a combination of cryogenic-electron microscopy (cryo-EM) single-particle analysis, computational structure prediction and biochemistry to reveal an intricate interaction network. A short linear motif in the mPSF subunit FIP1 interacts with the structured core of human PAPOA, with a binding mode that is evolutionary conserved from yeast to human. In higher eukaryotes, however, PAPOA contains a conserved C-terminal motif that can interact intramolecularly with the same residues of the PAPOA structured core used to bind FIP1. Interestingly, using biochemical assay and cryo-EM structural analysis, we found that the PAPOA C-terminal motif can also directly interact with mPSF at the subunit CPSF160. These results show that PAPOA recruitment to mPSF is mediated by two distinct intermolecular connections and further suggest the presence of mutually exclusive interactions in the regulation of 3' end processing. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19008.map.gz emd_19008.map.gz | 43 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19008-v30.xml emd-19008-v30.xml emd-19008.xml emd-19008.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19008.png emd_19008.png | 73.6 KB | ||
| Filedesc metadata |  emd-19008.cif.gz emd-19008.cif.gz | 7.3 KB | ||
| Others |  emd_19008_additional_1.map.gz emd_19008_additional_1.map.gz emd_19008_additional_2.map.gz emd_19008_additional_2.map.gz emd_19008_half_map_1.map.gz emd_19008_half_map_1.map.gz emd_19008_half_map_2.map.gz emd_19008_half_map_2.map.gz | 80.6 MB 76.1 MB 79.5 MB 79.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19008 http://ftp.pdbj.org/pub/emdb/structures/EMD-19008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19008 | HTTPS FTP |
-Related structure data
| Related structure data |  8r8rMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19008.map.gz / Format: CCP4 / Size: 85.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19008.map.gz / Format: CCP4 / Size: 85.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
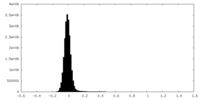
-Additional map: sharpened map - B factor 116.4
| File | emd_19008_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map - B factor 116.4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
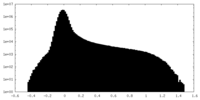
-Additional map: deepEMhancer map
| File | emd_19008_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
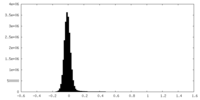
-Half map: #2
| File | emd_19008_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
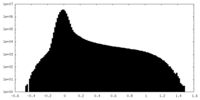
-Half map: #1
| File | emd_19008_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mPSF with PAPOA C-terminus peptide (PAPOAc)
| Entire | Name: mPSF with PAPOA C-terminus peptide (PAPOAc) |
|---|---|
| Components |
|
-Supramolecule #1: mPSF with PAPOA C-terminus peptide (PAPOAc)
| Supramolecule | Name: mPSF with PAPOA C-terminus peptide (PAPOAc) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cleavage and polyadenylation specificity factor subunit 1
| Macromolecule | Name: Cleavage and polyadenylation specificity factor subunit 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 161.074234 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYAVYKQAHP PTGLEFSMYC NFFNNSERNL VVAGTSQLYV YRLNRDAEAL TKNDRSTEGK AHREKLELAA SFSFFGNVMS MASVQLAGA KRDALLLSFK DAKLSVVEYD PGTHDLKTLS LHYFEEPELR DGFVQNVHTP RVRVDPDGRC AAMLVYGTRL V VLPFRRES ...String: MYAVYKQAHP PTGLEFSMYC NFFNNSERNL VVAGTSQLYV YRLNRDAEAL TKNDRSTEGK AHREKLELAA SFSFFGNVMS MASVQLAGA KRDALLLSFK DAKLSVVEYD PGTHDLKTLS LHYFEEPELR DGFVQNVHTP RVRVDPDGRC AAMLVYGTRL V VLPFRRES LAEEHEGLVG EGQRSSFLPS YIIDVRALDE KLLNIIDLQF LHGYYEPTLL ILFEPNQTWP GRVAVRQDTC SI VAISLNI TQKVHPVIWS LTSLPFDCTQ ALAVPKPIGG VVVFAVNSLL YLNQSVPPYG VALNSLTTGT TAFPLRTQEG VRI TLDCAQ ATFISYDKMV ISLKGGEIYV LTLITDGMRS VRAFHFDKAA ASVLTTSMVT MEPGYLFLGS RLGNSLLLKY TEKL QEPPA SAVREAADKE EPPSKKKRVD ATAGWSAAGK SVPQDEVDEI EVYGSEAQSG TQLATYSFEV CDSILNIGPC ANAAV GEPA FLSEEFQNSP EPDLEIVVCS GHGKNGALSV LQKSIRPQVV TTFELPGCYD MWTVIAPVRK EEEDNPKGEG TEQEPS TTP EADDDGRRHG FLILSREDST MILQTGQEIM ELDTSGFATQ GPTVFAGNIG DNRYIVQVSP LGIRLLEGVN QLHFIPV DL GAPIVQCAVA DPYVVIMSAE GHVTMFLLKS DSYGGRHHRL ALHKPPLHHQ SKVITLCLYR DLSGMFTTES RLGGARDE L GGRSGPEAEG LGSETSPTVD DEEEMLYGDS GSLFSPSKEE ARRSSQPPAD RDPAPFRAEP THWCLLVREN GTMEIYQLP DWRLVFLVKN FPVGQRVLVD SSFGQPTTQG EARREEATRQ GELPLVKEVL LVALGSRQSR PYLLVHVDQE LLIYEAFPHD SQLGQGNLK VRFKKVPHNI NFREKKPKPS KKKAEGGGAE EGAGARGRVA RFRYFEDIYG YSGVFICGPS PHWLLVTGRG A LRLHPMAI DGPVDSFAPF HNVNCPRGFL YFNRQGELRI SVLPAYLSYD APWPVRKIPL RCTAHYVAYH VESKVYAVAT ST NTPCARI PRMTGEEKEF ETIERDERYI HPQQEAFSIQ LISPVSWEAI PNARIELQEW EHVTCMKTVS LRSEETVSGL KGY VAAGTC LMQGEEVTCR GRILIMDVIE VVPEPGQPLT KNKFKVLYEK EQKGPVTALC HCNGHLVSAI GQKIFLWSLR ASEL TGMAF IDTQLYIHQM ISVKNFILAA DVMKSISLLR YQEESKTLSL VSRDAKPLEV YSVDFMVDNA QLGFLVSDRD RNLMV YMYL PEAKESFGGM RLLRRADFHV GAHVNTFWRT PCRGATEGLS KKSVVWENKH ITWFATLDGG IGLLLPMQEK TYRRLL MLQ NALTTMLPHH AGLNPRAFRM LHVDRRTLQN AVRNVLDGEL LNRYLYLSTM ERSELAKKIG TTPDIILDDL LETDRVT AH F UniProtKB:  Cleavage and polyadenylation specificity factor subunit 1 Cleavage and polyadenylation specificity factor subunit 1 |
-Macromolecule #2: pre-mRNA 3' end processing protein WDR33
| Macromolecule | Name: pre-mRNA 3' end processing protein WDR33 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.580129 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATEIGSPPR FFHMPRFQHQ APRQLFYKRP DFAQQQAMQQ LTFDGKRMRK AVNRKTIDYN PSVIKYLENR IWQRDQRDMR AIQPDAGYY NDLVPPIGML NNPMNAVTTK FVRTSTNKVK CPVFVVRWTP EGRRLVTGAS SGEFTLWNGL TFNFETILQA H DSPVRAMT ...String: MATEIGSPPR FFHMPRFQHQ APRQLFYKRP DFAQQQAMQQ LTFDGKRMRK AVNRKTIDYN PSVIKYLENR IWQRDQRDMR AIQPDAGYY NDLVPPIGML NNPMNAVTTK FVRTSTNKVK CPVFVVRWTP EGRRLVTGAS SGEFTLWNGL TFNFETILQA H DSPVRAMT WSHNDMWMLT ADHGGYVKYW QSNMNNVKMF QAHKEAIREA SFSPTDNKFA TCSDDGTVRI WDFLRCHEER IL RGHGADV KCVDWHPTKG LVVSGSKDSQ QPIKFWDPKT GQSLATLHAH KNTVMEVKLN LNGNWLLTAS RDHLCKLFDI RNL KEELQV FRGHKKEATA VAWHPVHEGL FASGGSDGSL LFWHVGVEKE VGGMEMAHEG MIWSLAWHPL GHILCSGSND HTSK FWTRN RPGDKMRD UniProtKB: pre-mRNA 3' end processing protein WDR33 |
-Macromolecule #3: Cleavage and polyadenylation specificity factor subunit 4
| Macromolecule | Name: Cleavage and polyadenylation specificity factor subunit 4 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.733426 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSAWSHPQFE KGGGSGGGSG GSAWSHPQFE KTAGLEVLFQ GPQEIIASVD HIKFDLEIAV EQQLGAQPLP FPGMDKSGAA VCEFFLKAA CGKGGMCPFR HISGEKTVVC KHWLRGLCKK GDQCEFLHEY DMTKMPECYF YSKFGECSNK ECPFLHIDPE S KIKDCPWY ...String: MSAWSHPQFE KGGGSGGGSG GSAWSHPQFE KTAGLEVLFQ GPQEIIASVD HIKFDLEIAV EQQLGAQPLP FPGMDKSGAA VCEFFLKAA CGKGGMCPFR HISGEKTVVC KHWLRGLCKK GDQCEFLHEY DMTKMPECYF YSKFGECSNK ECPFLHIDPE S KIKDCPWY DRGFCKHGPL CRHRHTRRVI CVNYLVGFCP EGPSCKFMHP RFELPMGTTE QPPLPQQTQP PAKQRTPQVI GV MQSQNSS AGNRGPRPLE QVTCYKCGEK GHYANRCTKG HLAFLSGQ UniProtKB:  Cleavage and polyadenylation specificity factor subunit 4 Cleavage and polyadenylation specificity factor subunit 4 |
-Macromolecule #4: cDNA FLJ50397, highly similar to Poly(A) polymerase alpha
| Macromolecule | Name: cDNA FLJ50397, highly similar to Poly(A) polymerase alpha type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.87241 KDa |
| Sequence | String: DLSDIPALPA NPIPVIKNSI KLRLNR UniProtKB: cDNA FLJ50397, highly similar to Poly(A) polymerase alpha |
-Macromolecule #5: RNA (5'-R(P*AP*AP*UP*AP*AP*A)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*AP*UP*AP*AP*A)-3') / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.907237 KDa |
| Sequence | String: AAUAAA |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.79 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 186241 |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X