+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | C. elegans TIR-1 protein. | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  SARM1 / TIR-1 / SARM1 / TIR-1 /  C. elegans / C. elegans /  neurodegeneration / neurodegeneration /  cryo-EM / cryo-EM /  structural biology / NAD+ metabolism / axon Wallerian degeneration / structural biology / NAD+ metabolism / axon Wallerian degeneration /  HYDROLASE HYDROLASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationserotonin biosynthetic process / negative regulation of MyD88-independent toll-like receptor signaling pathway / NAD catabolic process /  NAD+ nucleosidase activity / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / NAD+ nucleotidase, cyclic ADP-ribose generating / NADP+ nucleosidase activity / cell fate specification / response to axon injury / defense response to fungus ...serotonin biosynthetic process / negative regulation of MyD88-independent toll-like receptor signaling pathway / NAD catabolic process / NAD+ nucleosidase activity / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / NAD+ nucleotidase, cyclic ADP-ribose generating / NADP+ nucleosidase activity / cell fate specification / response to axon injury / defense response to fungus ...serotonin biosynthetic process / negative regulation of MyD88-independent toll-like receptor signaling pathway / NAD catabolic process /  NAD+ nucleosidase activity / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / NAD+ nucleotidase, cyclic ADP-ribose generating / NADP+ nucleosidase activity / cell fate specification / response to axon injury / defense response to fungus / signaling adaptor activity / axon cytoplasm / NAD+ nucleosidase activity / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / NAD+ nucleotidase, cyclic ADP-ribose generating / NADP+ nucleosidase activity / cell fate specification / response to axon injury / defense response to fungus / signaling adaptor activity / axon cytoplasm /  protein localization / protein localization /  small GTPase binding / cell-cell signaling / small GTPase binding / cell-cell signaling /  cell body / cell body /  nervous system development / defense response to Gram-negative bacterium / defense response to bacterium / nervous system development / defense response to Gram-negative bacterium / defense response to bacterium /  axon / negative regulation of gene expression / axon / negative regulation of gene expression /  innate immune response / innate immune response /  dendrite / positive regulation of gene expression / dendrite / positive regulation of gene expression /  protein kinase binding / protein kinase binding /  signal transduction / positive regulation of transcription by RNA polymerase II / identical protein binding signal transduction / positive regulation of transcription by RNA polymerase II / identical protein bindingSimilarity search - Function | ||||||||||||
| Biological species |   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) | ||||||||||||
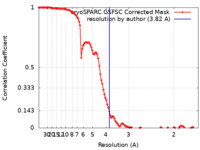
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.82 Å cryo EM / Resolution: 3.82 Å | ||||||||||||
 Authors Authors | Isupov MN / Opatowsky Y | ||||||||||||
| Funding support |  Israel, Israel,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans. Authors: Tami Khazma / Atira Grossman / Julia Guez-Haddad / Chengye Feng / Hadas Dabas / Radhika Sain / Michal Weitman / Ran Zalk / Michail N Isupov / Marc Hammarlund / Michael Hons / Yarden Opatowsky /     Abstract: Wallerian axonal degeneration (WD) does not occur in the nematode C. elegans, in contrast to other model animals. However, WD depends on the NADase activity of SARM1, a protein that is also expressed ...Wallerian axonal degeneration (WD) does not occur in the nematode C. elegans, in contrast to other model animals. However, WD depends on the NADase activity of SARM1, a protein that is also expressed in C. elegans (ceSARM/ceTIR-1). We hypothesized that differences in SARM between species might exist and account for the divergence in WD. We first show that expression of the human (h)SARM1, but not ceTIR-1, in C. elegans neurons is sufficient to confer axon degeneration after nerve injury. Next, we determined the cryoelectron microscopy structure of ceTIR-1 and found that, unlike hSARM1, which exists as an auto-inhibited ring octamer, ceTIR-1 forms a readily active 9-mer. Enzymatically, the NADase activity of ceTIR-1 is substantially weaker (10-fold higher Km) than that of hSARM1, and even when fully active, it falls short of consuming all cellular NAD. Our experiments provide insight into the molecular mechanisms and evolution of SARM orthologs and WD across species. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17370.map.gz emd_17370.map.gz | 61.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17370-v30.xml emd-17370-v30.xml emd-17370.xml emd-17370.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17370_fsc.xml emd_17370_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_17370.png emd_17370.png | 76 KB | ||
| Others |  emd_17370_half_map_1.map.gz emd_17370_half_map_1.map.gz emd_17370_half_map_2.map.gz emd_17370_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17370 http://ftp.pdbj.org/pub/emdb/structures/EMD-17370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17370 | HTTPS FTP |
-Related structure data
| Related structure data |  8p2mMC  8p2lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17370.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17370.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17370_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17370_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : C. elegans TIR1.
| Entire | Name: C. elegans TIR1. |
|---|---|
| Components |
|
-Supramolecule #1: C. elegans TIR1.
| Supramolecule | Name: C. elegans TIR1. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) |
-Macromolecule #1: NAD(+) hydrolase tir-1
| Macromolecule | Name: NAD(+) hydrolase tir-1 / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO / EC number: ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase |
|---|---|
| Source (natural) | Organism:   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) |
| Molecular weight | Theoretical: 84.496172 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSYHHHHHHD YDIPTTENLY FQGAMGSHMD ELSPVDQRST SGTARFLIQQ DSVVNPSTKM SNTEQVAMMH TLKTKLSKYQ AMMDKAFEE IAKVEDANII EGCTIVRKLM RKVWNTPKVS ADLANALCDY LRDRDYFDKL IKMFISPNTA ACDQVRMECG K VLEECTSS ...String: MSYHHHHHHD YDIPTTENLY FQGAMGSHMD ELSPVDQRST SGTARFLIQQ DSVVNPSTKM SNTEQVAMMH TLKTKLSKYQ AMMDKAFEE IAKVEDANII EGCTIVRKLM RKVWNTPKVS ADLANALCDY LRDRDYFDKL IKMFISPNTA ACDQVRMECG K VLEECTSS ANLEYIVNKS YTKKIMIVAM KLNKTPDQQR LSLSLIGNLF KHSNAVSLSL IETDVIDHII LTFKRAPECP DI LRHAALA LANILLYTCF EGKKKIIQKK IPEWLFFLAS QADDVTRYYA CIAVCTIVSV KEFEPLVRKS DTMKLVEPFL QVH DPATFA RDYHKYAQGN TKEWLERLLP MLQPSRRREA RSVAAFHFTL EATIKKEQNK LDVFQEIGAI QALKEVASSP DEVA AKFAS EALTVIGEEV PYKLAQQVPG WTCADVQYWV KKIGFEEYVE KFAKQMVDGD LLLQLTENDL KHDVGMISGL HRKRF LREL QTLKVAADYS SVDESNLDNF LMGLSPELSV YTYQMLTNGV NRSLLSSLTD EMMQNACGIT NPIHRLKLTQ AFETAK HPD DVEVAMLSKQ IDVFISYRRS TGNQLASLIK VLLQLRGYRV FIDVDKLYAG KFDSSLLKNI QAAKHFILVL TPNSLDR LL NDDNCEDWVH KELKCAFEHQ KNIIPIFDTA FEFPTKEDQI PNDIRMITKY NGVKWVHDYQ DACMAKVVRF ITGELNRT T PTTKEMPSIS RKTTQQR UniProtKB: NAD(+) hydrolase tir-1 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.8 µm |
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 41.4 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8p2m: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)