+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse Elp123 with bound SAM | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphorylase kinase regulator activity / tRNA uridine(34) acetyltransferase activity / elongator holoenzyme complex / tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation / I-kappaB phosphorylation / tRNA wobble uridine modification / regulation of receptor signaling pathway via JAK-STAT / RNA polymerase II complex binding /  acetyltransferase activity / endopeptidase activator activity ...phosphorylase kinase regulator activity / tRNA uridine(34) acetyltransferase activity / elongator holoenzyme complex / tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation / I-kappaB phosphorylation / tRNA wobble uridine modification / regulation of receptor signaling pathway via JAK-STAT / RNA polymerase II complex binding / acetyltransferase activity / endopeptidase activator activity ...phosphorylase kinase regulator activity / tRNA uridine(34) acetyltransferase activity / elongator holoenzyme complex / tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation / I-kappaB phosphorylation / tRNA wobble uridine modification / regulation of receptor signaling pathway via JAK-STAT / RNA polymerase II complex binding /  acetyltransferase activity / endopeptidase activator activity / acetyltransferase activity / endopeptidase activator activity /  proteasome assembly / proteasome assembly /  Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups /  proteasome complex / transcription elongation factor complex / proteasome complex / transcription elongation factor complex /  central nervous system development / transcription elongation by RNA polymerase II / central nervous system development / transcription elongation by RNA polymerase II /  neuron migration / neuron migration /  : / 4 iron, 4 sulfur cluster binding / : / 4 iron, 4 sulfur cluster binding /  tRNA binding / tRNA binding /  protein kinase activity / positive regulation of cell migration / regulation of transcription by RNA polymerase II / protein kinase activity / positive regulation of cell migration / regulation of transcription by RNA polymerase II /  nucleolus / nucleolus /  protein kinase binding / protein kinase binding /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.01 Å cryo EM / Resolution: 4.01 Å | |||||||||
 Authors Authors | Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D ...Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D / Lin T-Y / Abbassi N / Hammermeister A / Chramiec-Glabik A / Kosinski J / Schaffrath R / Glatt S | |||||||||
| Funding support |  Poland, European Union, 2 items Poland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Cryo-EM structure of the fully assembled Elongator complex. Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej ...Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej Chramiec-Głąbik / Anna Biela / Dominika Dobosz / Ting-Yu Lin / Nour-El-Hana Abbassi / Alexander Hammermeister / Juri Rappsilber / Jan Kosinski / Raffael Schaffrath / Sebastian Glatt /    Abstract: Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition ...Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition of chemical groups. Modifications in the tRNA anticodons are directly influencing ribosome decoding and dynamics during translation elongation and are crucial for maintaining proteome integrity. In eukaryotes, wobble uridines are modified by Elongator, a large and highly conserved macromolecular complex. Elongator consists of two subcomplexes, namely Elp123 containing the enzymatically active Elp3 subunit and the associated Elp456 hetero-hexamer. The structure of the fully assembled complex and the function of the Elp456 subcomplex have remained elusive. Here, we show the cryo-electron microscopy structure of yeast Elongator at an overall resolution of 4.3 Å. We validate the obtained structure by complementary mutational analyses in vitro and in vivo. In addition, we determined various structures of the murine Elongator complex, including the fully assembled mouse Elongator complex at 5.9 Å resolution. Our results confirm the structural conservation of Elongator and its intermediates among eukaryotes. Furthermore, we complement our analyses with the biochemical characterization of the assembled human Elongator. Our results provide the molecular basis for the assembly of Elongator and its tRNA modification activity in eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15682.map.gz emd_15682.map.gz | 15.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15682-v30.xml emd-15682-v30.xml emd-15682.xml emd-15682.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15682_fsc.xml emd_15682_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_15682.png emd_15682.png | 89.6 KB | ||
| Others |  emd_15682_half_map_1.map.gz emd_15682_half_map_1.map.gz emd_15682_half_map_2.map.gz emd_15682_half_map_2.map.gz | 226.5 MB 225.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15682 http://ftp.pdbj.org/pub/emdb/structures/EMD-15682 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15682 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15682 | HTTPS FTP |
-Related structure data
| Related structure data |  8avgMC  8asvC  8aswC  8at6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15682.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15682.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15682_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15682_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse Elp123 with bound SAM
| Entire | Name: Mouse Elp123 with bound SAM |
|---|---|
| Components |
|
-Supramolecule #1: Mouse Elp123 with bound SAM
| Supramolecule | Name: Mouse Elp123 with bound SAM / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 610 KDa |
-Macromolecule #1: Elongator complex protein 1
| Macromolecule | Name: Elongator complex protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 149.756922 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MRNLKLHRTL EFRDIQAPGK PQCFCLRAEQ GTVLIGSERG LTEVDPVRRE VKTEISLVAE GFLPEDGSGC IVGIQDLLDQ ESVCVATAS GDVIVCNLST QQLECVGSVA SGISVMSWSP DQELLLLATA QQTLIMMTKD FEVIAEEQIH QDDFGEGKFV T VGWGSKQT ...String: MRNLKLHRTL EFRDIQAPGK PQCFCLRAEQ GTVLIGSERG LTEVDPVRRE VKTEISLVAE GFLPEDGSGC IVGIQDLLDQ ESVCVATAS GDVIVCNLST QQLECVGSVA SGISVMSWSP DQELLLLATA QQTLIMMTKD FEVIAEEQIH QDDFGEGKFV T VGWGSKQT QFHGSEGRPT AFPVQLPENA LPWDDRRPHI TWRGDGQYFA VSVVCRQTEA RKIRVWNREF ALQSTSESVP GL GPALAWK PSGSLIASTQ DKPNQQDVVF FEKNGLLHGH FTLPFLKDEV KVNDLLWNAD SSVLAIWLED LPKEDSSTLK SYV QLWTVG NYHWYLKQSL PFSTTGKNQI VSLLWDPVTP CRLHVLCTGW RYLCCDWHWT TDRSSGNSAN DLANVAVIDG NRVL VTVFR QTVVPPPMCT YRLLIPHPVN QVIFSAHLGN DLAVLDASNQ ISVYKCGDKP NMDSTVKLGA VGGNGFKVPL TTPHL EKRY SIQFGNNEEE EEEEVNALQL SFLTWVEDDT FLAISYSHSS SQSIIHHLTV THSEVDEEQG QLDVSSSVTV DGVVIG LCC CSKTKSLAVQ LADGQVLKYL WESPSLAVEP WKNSEGIPVR FVHPCTQMEV ATIGGEECVL GLTDRCRFFI NDTEVAS NI TSFAVCDDFL LVTTHSHTCQ VFSLSGASLK MLQAALSGSH EASGEILRKV ERGSRIVTVV PQDTKLILQM PRGNLEVV H HRALVLAQIR KWLDKLMFKE AFECMRKLRI NLNLIHDHNP KVFLENVETF VKQIDSVNHI NLFFTELREE DVTKTMYPP PITKSVQVST HPDGKKLDLI CDAMRAAMEA INPRKFCLSI LTSHVKKTTP ELEIVLQKVQ ELQGNLPFDP ESVSVEEALK YLLLLVDVN ELFNHSLGTY DFNLVLMVAE KSQKDPKEYL PFLNTLKKME TNYQRFTIDK YLKRYEKALG HLSKCGPEYF T ECLNLIKD KNLYKEALKL YRPDSPQYQA VSMAYGEHLM QEHLYEPAGL VFARCGAQEK ALEAFLACGS WQQALCVAAQ LQ MSKDKVA GLARTLAGKL VEQRKHSEAA TVLEQYAQDY EEAVLLLLEG SAWEEALRLV YKYDRVDIIE TSVKPSILEA QKN YMDFLD SETATFIRHK NRLQVVRALR RQAPQVHVDH EVAHGPESDL FSETSSIMSG SEMSGRYSHS NSRISARSSK NRRK AERKK HSLKEGSPLE GLALLEALSE VVQSVEKLKD EVRAILKVLF LFEFEEQAKE LQRAFESTLQ LMERAVPEIW TPAGQ QSST TPVLGPSSTA NSITASYQQQ KTCVPALDAG VYMPPKMDPR SQWKLSLLE |
-Macromolecule #2: Elongator complex protein 2
| Macromolecule | Name: Elongator complex protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 93.193773 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MVSSVLEVSH VFCCPNRVRG ALSWNTGPGG LLAFGTSCSV VLYDPQKKVV ITNLNGHTAR VNCLQWIRTE DGSPSNELVS GGSDNRVIH WELENNQVLK SVRLQGHEGP VCAVHAIYQS GPSEGEQHAL IASAASDSTV RIWSKKGSEV KYLQTLSFRD G FVLSVCLA ...String: MVSSVLEVSH VFCCPNRVRG ALSWNTGPGG LLAFGTSCSV VLYDPQKKVV ITNLNGHTAR VNCLQWIRTE DGSPSNELVS GGSDNRVIH WELENNQVLK SVRLQGHEGP VCAVHAIYQS GPSEGEQHAL IASAASDSTV RIWSKKGSEV KYLQTLSFRD G FVLSVCLA ILPGTNVPVL ACGDDDCRIH LYIQQDDQFQ KALSLCGHED WIRGVEWATF GRDLFLASCS QDCLIRIWRL YM KPASFET KDGSLRLKEN TFTIKDGGVR TTVAVTLETV LAGHENWVNA VHWQPSFYKD GVLQQPVRLL SASMDKTMIL WAP DEESGV WLEQVRVGEV GGNTLGFYDC QFGENGTMII AHAFHGALHL WKQSTVNPRQ WAPEIVISGH FDGVQDLMWD PEGE FIITT STDQTTRLFA PWKKKDQKDR SQVTWHEIAR PQIHGYNIKC LAMIDRFQFV SGADEKVLRV FSAPRNFVEN FSVIS RQSL SHMLCDDQDL PEGATVPALG LSNKALFQGD IASQPFEEDE LISPAFGSPQ VTFQPAVLNE PPTEDHLLQN TLWPEI QKL YGHGYEIVCV ACNNSKTLLA SACKASQKEH AAIILWSTAS WKQVQSLAFH TLTVTQMTFS PDDKFLLAVS RDRTWSL WK RQDATSSEFD PFFSLFAFTN KITSVHSRII WSCDWSPDNK YFFTGSRDKK VVVWGECKSS HNPMEHPIRP CSSILDVG S SVTAVSVCPV LNPAQRYIVA IGLESGKICI YSWNKTNQEI NDWTSCVETN PSQSHSLGIR RLCWKSCSDD DDDDDDDDT EQSEEGPEWL HFASCGEDHT VKIYRVNRRA L |
-Macromolecule #3: Elongator complex protein 3
| Macromolecule | Name: Elongator complex protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number:  Transferases; Acyltransferases; Transferring groups other than aminoacyl groups Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 62.481059 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MRQKRKGDLS PAELMMLTIG DVIKQLVEAH EQGKDVDLNK MKTKTAAKYG LASQPRLVDI IAAVPPHYRK ILIPKLKAKP VRTASGIAV VAVMCKPHRC PHISFTGNIC IYCPGGPDSD FEYSTQSYTG YEPTSMRAIR ARYDPFLQTR HRIEQLKQLG H SVDKVEFI ...String: MRQKRKGDLS PAELMMLTIG DVIKQLVEAH EQGKDVDLNK MKTKTAAKYG LASQPRLVDI IAAVPPHYRK ILIPKLKAKP VRTASGIAV VAVMCKPHRC PHISFTGNIC IYCPGGPDSD FEYSTQSYTG YEPTSMRAIR ARYDPFLQTR HRIEQLKQLG H SVDKVEFI VMGGTFMALP EEYRDYFIRS LHDALSGHTS NNIHEAIKYS ERSFTKCVGI TIETRPDYCM KRHLSDMLTY GC TRLEIGV QSVYEDVARD TNRGHTVKAA CESFHLAKDS GFKVVTHMMP DLPNVGLERD IEQFIEFFEN PAFRPDGLKL YPT LVIRGT GLYELWKSGR YRSYSPSDLI ELVARILALV PPWTRVYRVQ RDIPMPLVSS GVEHGNLREL AFARMKDLGI QCRD VRTRE VGIQEIHHRV RPYQVELVRR DYVANGGWET FLSYEDPDQD ILIGLLRLRK CSEETFRFEL GGGVSIVREL HVYGS VVPV SSRDPTKFQH QGFGMLLMEE AERIAREEHG SGKMAVISGV GTRNYYRKIG YRLQGPYMVK MLK |
-Macromolecule #4: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 4 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #5: S-ADENOSYLMETHIONINE
| Macromolecule | Name: S-ADENOSYLMETHIONINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: SAM |
|---|---|
| Molecular weight | Theoretical: 398.437 Da |
| Chemical component information | 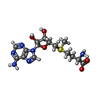 ChemComp-SAM: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 15 s wait time, blot force 5, 5 s blot time. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 6415 / Average exposure time: 1.82 sec. / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






































