[English] 日本語
 Yorodumi
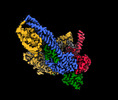
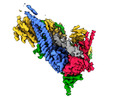
Yorodumi- EMDB-13787: Cryo-EM map of clamped S.cerevisiae condensin complex on circular DNA -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of clamped S.cerevisiae condensin complex on circular DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  condensin / SMC / condensin / SMC /  CELL CYCLE CELL CYCLE | |||||||||
| Biological species |   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 8.75 Å cryo EM / Resolution: 8.75 Å | |||||||||
 Authors Authors | Lee B-G / Rhodes J / Lowe J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Clamping of DNA shuts the condensin neck gate. Authors: Byung-Gil Lee / James Rhodes / Jan Löwe /  Abstract: SignificanceDNA needs to be compacted to fit into nuclei and during cell division, when dense chromatids are formed for their mechanical segregation, a process that depends on the protein complex ...SignificanceDNA needs to be compacted to fit into nuclei and during cell division, when dense chromatids are formed for their mechanical segregation, a process that depends on the protein complex condensin. It forms and enlarges loops in DNA through loop extrusion. Our work resolves the atomic structure of a DNA-bound state of condensin in which ATP has not been hydrolyzed. The DNA is clamped within a compartment that has been reported previously in other structural maintenance of chromosomes (SMC) complexes, including Rad50, cohesin, and MukBEF. With the caveat of important differences, it means that all SMC complexes cycle through at least some similar states and undergo similar conformational changes in their head modules, while hydrolyzing ATP and translocating DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13787.map.gz emd_13787.map.gz | 15.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13787-v30.xml emd-13787-v30.xml emd-13787.xml emd-13787.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13787.png emd_13787.png | 58 KB | ||
| Filedesc metadata |  emd-13787.cif.gz emd-13787.cif.gz | 3.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13787 http://ftp.pdbj.org/pub/emdb/structures/EMD-13787 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13787 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13787 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13787.map.gz / Format: CCP4 / Size: 16.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13787.map.gz / Format: CCP4 / Size: 16.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : complex of condensin tetraner (smc2, smc4, brn1, ycs4), ADP-BeF3 ...
| Entire | Name: complex of condensin tetraner (smc2, smc4, brn1, ycs4), ADP-BeF3 on circular DNA |
|---|---|
| Components |
|
-Supramolecule #1: complex of condensin tetraner (smc2, smc4, brn1, ycs4), ADP-BeF3 ...
| Supramolecule | Name: complex of condensin tetraner (smc2, smc4, brn1, ycs4), ADP-BeF3 on circular DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae S288C (yeast) Saccharomyces cerevisiae S288C (yeast) |
| Molecular weight | Theoretical: 515 kDa/nm |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 32.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: cryoSPARC |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: cryoSPARC |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 8.75 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 36588 |
 Movie
Movie Controller
Controller