[English] 日本語
 Yorodumi
Yorodumi- EMDB-13657: Structure of double-stranded DNA-bound MCM2-7 DH complexed with C... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of double-stranded DNA-bound MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATP (B1 map) | |||||||||||||||
 Map data Map data | Body 1 of multi-body refinement of MD-(ATP) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords |  Helicase / Helicase /  Activation / Activation /  Kinase / Kinase /  Phosphorylation / Phosphorylation /  REPLICATION REPLICATION | |||||||||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||||||||
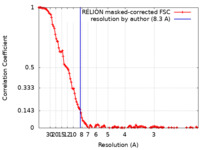
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 8.3 Å cryo EM / Resolution: 8.3 Å | |||||||||||||||
 Authors Authors | Saleh A / Noguchi Y / Aramayo R / Ivanova ME / Speck C | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer. Authors: Almutasem Saleh / Yasunori Noguchi / Ricardo Aramayo / Marina E Ivanova / Kathryn M Stevens / Alex Montoya / S Sunidhi / Nicolas Lopez Carranza / Marcin J Skwark / Christian Speck /  Abstract: The controlled assembly of replication forks is critical for genome stability. The Dbf4-dependent Cdc7 kinase (DDK) initiates replisome assembly by phosphorylating the MCM2-7 replicative helicase at ...The controlled assembly of replication forks is critical for genome stability. The Dbf4-dependent Cdc7 kinase (DDK) initiates replisome assembly by phosphorylating the MCM2-7 replicative helicase at the N-terminal tails of Mcm2, Mcm4 and Mcm6. At present, it remains poorly understood how DDK docks onto the helicase and how the kinase targets distal Mcm subunits for phosphorylation. Using cryo-electron microscopy and biochemical analysis we discovered that an interaction between the HBRCT domain of Dbf4 with Mcm2 serves as an anchoring point, which supports binding of DDK across the MCM2-7 double-hexamer interface and phosphorylation of Mcm4 on the opposite hexamer. Moreover, a rotation of DDK along its anchoring point allows phosphorylation of Mcm2 and Mcm6. In summary, our work provides fundamental insights into DDK structure, control and selective activation of the MCM2-7 helicase during DNA replication. Importantly, these insights can be exploited for development of novel DDK inhibitors. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13657.map.gz emd_13657.map.gz | 138.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13657-v30.xml emd-13657-v30.xml emd-13657.xml emd-13657.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13657_fsc.xml emd_13657_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_13657.png emd_13657.png | 48.8 KB | ||
| Masks |  emd_13657_msk_1.map emd_13657_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13657.cif.gz emd-13657.cif.gz | 4.4 KB | ||
| Others |  emd_13657_additional_1.map.gz emd_13657_additional_1.map.gz | 19 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13657 http://ftp.pdbj.org/pub/emdb/structures/EMD-13657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13657 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13657.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13657.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body 1 of multi-body refinement of MD-(ATP) | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.085 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13657_msk_1.map emd_13657_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Post-processed map of Body 1 of multi-body refinement...
| File | emd_13657_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map of Body 1 of multi-body refinement of MD-(ATP) (B-factor of -100 applied) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MCM2-7 double hexamer bound to double-stranded DNA and two copies...
| Entire | Name: MCM2-7 double hexamer bound to double-stranded DNA and two copies of Cdc7-Dbf4 |
|---|---|
| Components |
|
-Supramolecule #1: MCM2-7 double hexamer bound to double-stranded DNA and two copies...
| Supramolecule | Name: MCM2-7 double hexamer bound to double-stranded DNA and two copies of Cdc7-Dbf4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#9 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Molecular weight | Theoretical: 1.54 MDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 1.5 seconds and blot force +2. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 75000 Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number real images: 3416 / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z
Z Y
Y X
X