+Search query
-Structure paper
| Title | The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 2915, Year 2022 |
| Publish date | May 25, 2022 |
 Authors Authors | Almutasem Saleh / Yasunori Noguchi / Ricardo Aramayo / Marina E Ivanova / Kathryn M Stevens / Alex Montoya / S Sunidhi / Nicolas Lopez Carranza / Marcin J Skwark / Christian Speck /  |
| PubMed Abstract | The controlled assembly of replication forks is critical for genome stability. The Dbf4-dependent Cdc7 kinase (DDK) initiates replisome assembly by phosphorylating the MCM2-7 replicative helicase at ...The controlled assembly of replication forks is critical for genome stability. The Dbf4-dependent Cdc7 kinase (DDK) initiates replisome assembly by phosphorylating the MCM2-7 replicative helicase at the N-terminal tails of Mcm2, Mcm4 and Mcm6. At present, it remains poorly understood how DDK docks onto the helicase and how the kinase targets distal Mcm subunits for phosphorylation. Using cryo-electron microscopy and biochemical analysis we discovered that an interaction between the HBRCT domain of Dbf4 with Mcm2 serves as an anchoring point, which supports binding of DDK across the MCM2-7 double-hexamer interface and phosphorylation of Mcm4 on the opposite hexamer. Moreover, a rotation of DDK along its anchoring point allows phosphorylation of Mcm2 and Mcm6. In summary, our work provides fundamental insights into DDK structure, control and selective activation of the MCM2-7 helicase during DNA replication. Importantly, these insights can be exploited for development of novel DDK inhibitors. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35614055 / PubMed:35614055 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.2 - 18.0 Å |
| Structure data | EMDB-13619: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (composite map) EMDB-13620, PDB-7pt7:  EMDB-13621: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (B1 map)  EMDB-13622: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (B2 map)  EMDB-13623: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (B3 map)  EMDB-13624: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state III (3D auto-refined map)  EMDB-13629: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (composite map)  EMDB-13631: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (B1 map)  EMDB-13635: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (B2 map)  EMDB-13640: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state II (3D auto-refined map)  EMDB-13644: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATPgS, state I (3D auto-refined map)  EMDB-13645: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, state I (B1 map)  EMDB-13646: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, state I (B2 map)  EMDB-13647: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, state I (3D auto-refined map)  EMDB-13648: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, swiveled state (3D auto-refined map)  EMDB-13649: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, swiveled state A (binned 3D auto-refined map)  EMDB-13650: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, swiveled state B (binned 3D auto-refined map)  EMDB-13651: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, swiveled state C (binned 3D auto-refined map)  EMDB-13652: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, swiveled state D (binned 3D auto-refined map)  EMDB-13653: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, swiveled state E (binned 3D auto-refined map)  EMDB-13655: Structure of MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ADP:BeF3, swiveled state F (binned 3D auto-refined map)  EMDB-13656: Structure of double-stranded DNA-bound MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATP (3D auto-refined map)  EMDB-13657: Structure of double-stranded DNA-bound MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATP (B1 map)  EMDB-13658: Structure of double-stranded DNA-bound MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATP (B2 map)  EMDB-13659: Structure of double-stranded DNA-bound MCM2-7 DH complexed with Cdc7-Dbf4 in the presence of ATP (B3 map) |
| Chemicals | 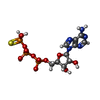 ChemComp-AGS:  ChemComp-MG:  ChemComp-ZN:  ChemComp-ADP: 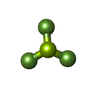 ChemComp-BEF: |
| Source |
|
 Keywords Keywords |  REPLICATION / REPLICATION /  Helicase / Helicase /  Activation / Activation /  Kinase / Kinase /  Phosphorylation Phosphorylation |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








