[English] 日本語
 Yorodumi
Yorodumi- EMDB-40979: Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 tri... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
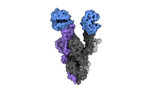
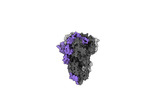
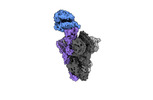
| Title | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation | |||||||||
 Map data Map data | Cryo-EM Map of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Coronavirus / Spike / Coronavirus / Spike /  ACE2 / Mink / ACE2 / Mink /  PROTEIN BINDING / PROTEIN BINDING /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity /  carboxypeptidase activity / carboxypeptidase activity /  cilium / cilium /  metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface ... metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface ... Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity /  carboxypeptidase activity / carboxypeptidase activity /  cilium / cilium /  metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  proteolysis / extracellular region / proteolysis / extracellular region /  membrane / identical protein binding / membrane / identical protein binding /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 / Severe acute respiratory syndrome coronavirus 2 /   Neovison vison (American mink) / Neovison vison (American mink) /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.87 Å cryo EM / Resolution: 3.87 Å | |||||||||
 Authors Authors | Ahn HM / Calderon B / Fan X / Gao Y / Horgan N / Zhou B / Liang B | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Med Virol / Year: 2023 Journal: J Med Virol / Year: 2023Title: Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2. Authors: Hyunjun Ahn / Brenda M Calderon / Xiaoyu Fan / Yunrong Gao / Natalie L Horgan / Nannan Jiang / Dylan S Blohm / Jaber Hossain / Nicole Wedad K Rayyan / Sarah H Osman / Xudong Lin / Michael ...Authors: Hyunjun Ahn / Brenda M Calderon / Xiaoyu Fan / Yunrong Gao / Natalie L Horgan / Nannan Jiang / Dylan S Blohm / Jaber Hossain / Nicole Wedad K Rayyan / Sarah H Osman / Xudong Lin / Michael Currier / John Steel / David E Wentworth / Bin Zhou / Bo Liang /  Abstract: Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) enters the host cell by binding to angiotensin-converting enzyme 2 (ACE2). While evolutionarily conserved, ACE2 receptors differ across ...Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) enters the host cell by binding to angiotensin-converting enzyme 2 (ACE2). While evolutionarily conserved, ACE2 receptors differ across various species and differential interactions with Spike (S) glycoproteins of SARS-CoV-2 viruses impact species specificity. Reverse zoonoses led to SARS-CoV-2 outbreaks on multiple American mink (Mustela vison) farms during the pandemic and gave rise to mink-associated S substitutions known for transmissibility between mink and zoonotic transmission to humans. In this study, we used bio-layer interferometry (BLI) to discern the differences in binding affinity between multiple human and mink-derived S glycoproteins of SARS-CoV-2 and their respective ACE2 receptors. Further, we conducted a structural analysis of a mink variant S glycoprotein and American mink ACE2 (mvACE2) using cryo-electron microscopy (cryo-EM), revealing four distinct conformations. We discovered a novel intermediary conformation where the mvACE2 receptor is bound to the receptor-binding domain (RBD) of the S glycoprotein in a "down" position, approximately 34° lower than previously reported "up" RBD. Finally, we compared residue interactions in the S-ACE2 complex interface of S glycoprotein conformations with varying RBD orientations. These findings provide valuable insights into the molecular mechanisms of SARS-CoV-2 entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40979.map.gz emd_40979.map.gz | 482.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40979-v30.xml emd-40979-v30.xml emd-40979.xml emd-40979.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40979_fsc.xml emd_40979_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40979.png emd_40979.png | 12 KB | ||
| Filedesc metadata |  emd-40979.cif.gz emd-40979.cif.gz | 6.6 KB | ||
| Others |  emd_40979_half_map_1.map.gz emd_40979_half_map_1.map.gz emd_40979_half_map_2.map.gz emd_40979_half_map_2.map.gz | 474.6 MB 474.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40979 http://ftp.pdbj.org/pub/emdb/structures/EMD-40979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40979 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40979 | HTTPS FTP |
-Related structure data
| Related structure data |  8t23MC  8t20C  8t21C  8t22C  8t25C  8tazC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40979.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40979.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM Map of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map of the density of the RBD-ACE2...
| File | emd_40979_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of the density of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of the density of the RBD-ACE2...
| File | emd_40979_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of the density of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mink variant spike RBD-ACE2 interface after local refinement
| Entire | Name: mink variant spike RBD-ACE2 interface after local refinement |
|---|---|
| Components |
|
-Supramolecule #1: mink variant spike RBD-ACE2 interface after local refinement
| Supramolecule | Name: mink variant spike RBD-ACE2 interface after local refinement type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Macromolecule #1: Angiotensin-converting enzyme
| Macromolecule | Name: Angiotensin-converting enzyme / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Neovison vison (American mink) Neovison vison (American mink) |
| Molecular weight | Theoretical: 89.3755 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLGSSWLLLS LAALTAAQST TEDLAKTFLE KFNYEAEELS YQNSLASWNY NTNITDENIQ KMNIAGAKWS AFYEEESQHA KTYPLEEIQ DPIIKRQLRA LQQSGSSVLS ADKRERLNTI LNAMSTIYST GKACNPNNPQ ECLLLEPGLD DIMENSKDYN E RLWAWEGW ...String: MLGSSWLLLS LAALTAAQST TEDLAKTFLE KFNYEAEELS YQNSLASWNY NTNITDENIQ KMNIAGAKWS AFYEEESQHA KTYPLEEIQ DPIIKRQLRA LQQSGSSVLS ADKRERLNTI LNAMSTIYST GKACNPNNPQ ECLLLEPGLD DIMENSKDYN E RLWAWEGW RSEVGKQLRP LYEEYVALKN EMARANNYED YGDYWRGDYE EEWADGYNYS RNQLIEDVEH TFTQIKPLYE HL HAYVRAK LMDAYPSRIS PTGCLPAHLL GDMWGRFWTN LYPLMVPFGQ KPNIDVTDAM VNQSWDARRI FKEAEKFFVS VGL PNMTEG FWQNSMLTEP GDNRKVVCHP TAWDLGKHDF RIKMCTKVTM DDFLTAHHEM GHIQYDMAYA AQPFLLRNGA NEGF HEAVG EIMSLSAATP NHLKNIGLLP PDFSEDSETD INFLLKQALT IVGTLPFTYM LEKWRWMVFK GEIPKEQWMQ KWWEM KRDI VGVVEPLPHD ETYCDPAALF HVANDYSFIR YYTRTIYQFQ FQEALCQIAK HEGPLYKCDI SNSREAGQKL HEMLSL GRS KPWTFALERV VGAKTMDVRP LLNYFEPLFT WLKEQNRNSF VGWNTDWSPY ADQSIKVRIS LKSALGEKAY EWNDNEM YF FQSSIAYAMR EYFSKVKKQT IPFVDKDVRV SDLKPRISFN FIVTSPENMS DIIPRADVEE AIRKSRGRIN DAFRLDDN S LEFLGIQPTL EPPYQPPVGS GSGSGHHHHH HGSGSGLNDI FEAQKIEWHE UniProtKB:  Angiotensin-converting enzyme Angiotensin-converting enzyme |
-Macromolecule #2: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.370246 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQ IAPGQTGKIA DYNYKLPDDF TGCVIAWNSN NLDSKVGGNY NYLFRLFRKS NLKPFERDIS TEIYQAGSTP C NGVEGFNC ...String: QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQ IAPGQTGKIA DYNYKLPDDF TGCVIAWNSN NLDSKVGGNY NYLFRLFRKS NLKPFERDIS TEIYQAGSTP C NGVEGFNC YFPLQSYGFQ PTNGVGYQPY RVVVLSFELL HAPATVCGPK UniProtKB:  Spike glycoprotein Spike glycoprotein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20 mM Tris pH 8.0, 200 mM NaCl | |||||||||
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 1.75 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 11392 / Average exposure time: 4.0 sec. / Average electron dose: 49.98 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X



















