[English] 日本語
 Yorodumi
Yorodumi- PDB-8taz: Cryo-EM structure of mink variant Y453F trimeric spike protein bo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8taz | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  VIRAL PROTEIN / VIRAL PROTEIN /  Coronavirus / Spike / Coronavirus / Spike /  ACE2 / Mink / ACE2 / Mink /  protein binding protein binding | ||||||
| Function / homology |  Function and homology information Function and homology information Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity /  carboxypeptidase activity / carboxypeptidase activity /  cilium / cilium /  metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface ... metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface ... Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity / Hydrolases; Acting on peptide bonds (peptidases) / peptidyl-dipeptidase activity /  carboxypeptidase activity / carboxypeptidase activity /  cilium / cilium /  metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / metallopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  proteolysis / extracellular region / proteolysis / extracellular region /  membrane / identical protein binding / membrane / identical protein binding /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2  Neogale vison (American mink) Neogale vison (American mink) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.75 Å cryo EM / Resolution: 3.75 Å | ||||||
 Authors Authors | Ahn, H.M. / Calderon, B. / Fan, X. / Gao, Y. / Horgan, N. / Liang, B. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: J Med Virol / Year: 2023 Journal: J Med Virol / Year: 2023Title: Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2. Authors: Hyunjun Ahn / Brenda M Calderon / Xiaoyu Fan / Yunrong Gao / Natalie L Horgan / Nannan Jiang / Dylan S Blohm / Jaber Hossain / Nicole Wedad K Rayyan / Sarah H Osman / Xudong Lin / Michael ...Authors: Hyunjun Ahn / Brenda M Calderon / Xiaoyu Fan / Yunrong Gao / Natalie L Horgan / Nannan Jiang / Dylan S Blohm / Jaber Hossain / Nicole Wedad K Rayyan / Sarah H Osman / Xudong Lin / Michael Currier / John Steel / David E Wentworth / Bin Zhou / Bo Liang /  Abstract: Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) enters the host cell by binding to angiotensin-converting enzyme 2 (ACE2). While evolutionarily conserved, ACE2 receptors differ across ...Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) enters the host cell by binding to angiotensin-converting enzyme 2 (ACE2). While evolutionarily conserved, ACE2 receptors differ across various species and differential interactions with Spike (S) glycoproteins of SARS-CoV-2 viruses impact species specificity. Reverse zoonoses led to SARS-CoV-2 outbreaks on multiple American mink (Mustela vison) farms during the pandemic and gave rise to mink-associated S substitutions known for transmissibility between mink and zoonotic transmission to humans. In this study, we used bio-layer interferometry (BLI) to discern the differences in binding affinity between multiple human and mink-derived S glycoproteins of SARS-CoV-2 and their respective ACE2 receptors. Further, we conducted a structural analysis of a mink variant S glycoprotein and American mink ACE2 (mvACE2) using cryo-electron microscopy (cryo-EM), revealing four distinct conformations. We discovered a novel intermediary conformation where the mvACE2 receptor is bound to the receptor-binding domain (RBD) of the S glycoprotein in a "down" position, approximately 34° lower than previously reported "up" RBD. Finally, we compared residue interactions in the S-ACE2 complex interface of S glycoprotein conformations with varying RBD orientations. These findings provide valuable insights into the molecular mechanisms of SARS-CoV-2 entry. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8taz.cif.gz 8taz.cif.gz | 765.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8taz.ent.gz pdb8taz.ent.gz | 517.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8taz.json.gz 8taz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ta/8taz https://data.pdbj.org/pub/pdb/validation_reports/ta/8taz ftp://data.pdbj.org/pub/pdb/validation_reports/ta/8taz ftp://data.pdbj.org/pub/pdb/validation_reports/ta/8taz | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 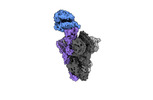 41143MC  8t20C  8t21C  8t22C  8t23C  8t25C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein |  Spike protein / S glycoprotein / E2 / Peplomer protein Spike protein / S glycoprotein / E2 / Peplomer proteinMass: 140244.109 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2Gene: S, 2 / Production host:   Homo sapiens (human) / References: UniProt: P0DTC2 Homo sapiens (human) / References: UniProt: P0DTC2#2: Protein | |  Mass: 89375.500 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Neogale vison (American mink) / Production host: Neogale vison (American mink) / Production host:   Homo sapiens (human) / References: UniProt: A0A7T0Q2W2 Homo sapiens (human) / References: UniProt: A0A7T0Q2W2 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors Type: COMPLEX / Entity ID: all / Source: MULTIPLE SOURCES | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| |||||||||||||||
| Source (recombinant) | Organism:   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Buffer solution | pH: 8 / Details: 20 mM Tris pH 8.0, 200 mM NaCl | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 1.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | |||||||||||||||
| Specimen support | Grid material: COPPER | |||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: OTHER / Nominal magnification: 81000 X / Nominal defocus max: 1750 nm / Nominal defocus min: 750 nm / C2 aperture diameter: 100 µm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 4 sec. / Electron dose: 49.98 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of real images: 11392 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.75 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 130988 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 221.77 Å2 | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj



