+Search query
-Structure paper
| Title | Structural basis of human Slo2.2 channel gating and modulation. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 42, Issue 8, Page 112858, Year 2023 |
| Publish date | Jul 25, 2023 |
 Authors Authors | Jiangtao Zhang / Shiqi Liu / Junping Fan / Rui Yan / Bo Huang / Feng Zhou / Tian Yuan / Jianke Gong / Zhuo Huang / Daohua Jiang /  |
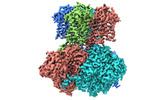
| PubMed Abstract | The sodium-activated Slo2.2 channel is abundantly expressed in the brain, playing a critical role in regulating neuronal excitability. The Na-binding site and the underlying mechanisms of Na- ...The sodium-activated Slo2.2 channel is abundantly expressed in the brain, playing a critical role in regulating neuronal excitability. The Na-binding site and the underlying mechanisms of Na-dependent activation remain unclear. Here, we present cryoelectron microscopy (cryo-EM) structures of human Slo2.2 in closed, open, and inhibitor-bound form at resolutions of 2.6-3.2 Å, revealing gating mechanisms of Slo2.2 regulation by cations and a potent inhibitor. The cytoplasmic gating ring domain of the closed Slo2.2 harbors multiple K and Zn sites, which stabilize the channel in the closed conformation. The open Slo2.2 structure reveals at least two Na-sensitive sites where Na binding induces expansion and rotation of the gating ring that opens the inner gate. Furthermore, a potent inhibitor wedges into a pocket formed by pore helix and S6 helix and blocks the pore. Together, our results provide a comprehensive structural framework for the investigation of Slo2.2 channel gating, Na sensation, and inhibition. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:37494189 PubMed:37494189 |
| Methods | EM (single particle) |
| Resolution | 2.64 - 3.18 Å |
| Structure data | EMDB-34827, PDB-8hir: EMDB-34847, PDB-8hk6: EMDB-34851, PDB-8hkf: EMDB-34853, PDB-8hkk: EMDB-34855, PDB-8hkm: EMDB-34858, PDB-8hkq: |
| Chemicals |  ChemComp-NA:  ChemComp-K: 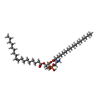 ChemComp-6OU:  ChemComp-ZN:  ChemComp-LQ9: |
| Source |
|
 Keywords Keywords |  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  potassium channels / potassium channels /  potassium channel / potassium channel /  ion channel ion channel |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers