+Search query
-Structure paper
| Title | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity. |
|---|---|
| Journal, issue, pages | Nat Chem Biol, Vol. 18, Issue 9, Page 972-980, Year 2022 |
| Publish date | Jun 23, 2022 |
 Authors Authors | Huasong Ai / Maoshen Sun / Aijun Liu / Zixian Sun / Tingting Liu / Lin Cao / Lujun Liang / Qian Qu / Zichen Li / Zhiheng Deng / Zebin Tong / Guochao Chu / Xiaolin Tian / Haiteng Deng / Suwen Zhao / Jia-Bin Li / Zhiyong Lou / Lei Liu /  |
| PubMed Abstract | Ubiquitination-dependent histone crosstalk plays critical roles in chromatin-associated processes and is highly associated with human diseases. Mechanism studies of the crosstalk have been of the ...Ubiquitination-dependent histone crosstalk plays critical roles in chromatin-associated processes and is highly associated with human diseases. Mechanism studies of the crosstalk have been of the central focus. Here our study on the crosstalk between H2BK34ub and Dot1L-catalyzed H3K79me suggests a novel mechanism of ubiquitination-induced nucleosome distortion to stimulate the activity of an enzyme. We determined the cryo-electron microscopy structures of Dot1L-H2BK34ub nucleosome complex and the H2BK34ub nucleosome alone. The structures reveal that H2BK34ub induces an almost identical orientation and binding pattern of Dot1L on nucleosome as H2BK120ub, which positions Dot1L for the productive conformation through direct ubiquitin-enzyme contacts. However, H2BK34-anchored ubiquitin does not directly interact with Dot1L as occurs in the case of H2BK120ub, but rather induces DNA and histone distortion around the modified site. Our findings establish the structural framework for understanding the H2BK34ub-H3K79me trans-crosstalk and highlight the diversity of mechanisms for histone ubiquitination to activate chromatin-modifying enzymes. |
 External links External links |  Nat Chem Biol / Nat Chem Biol /  PubMed:35739357 PubMed:35739357 |
| Methods | EM (single particle) |
| Resolution | 2.57 - 3.48 Å |
| Structure data | EMDB-33126, PDB-7xcr: EMDB-33127, PDB-7xct:  EMDB-33128: cryo-EM map of Dot1L and H2BK34ub-H3K79Nle nucleosome complex containing addition map (1:1) EMDB-33131, PDB-7xd0: EMDB-33132, PDB-7xd1:  EMDB-33139: cryo-EM map of Dot1L and H3K79Nle nucleosome complex (active state)  EMDB-33141: cryo-EM map of Dot1L and H3K79Nle nucleosome complex (inactive state) |
| Chemicals | 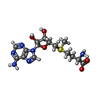 ChemComp-SAM: |
| Source |
|
 Keywords Keywords |  NUCLEAR PROTEIN / NUCLEAR PROTEIN /  Complex / Complex /  Dot1L / H2BK34ub / Dot1L / H2BK34ub /  Nucleosome / H2BK34ub nucleosome / unmodified nucleosome Nucleosome / H2BK34ub nucleosome / unmodified nucleosome |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers