+Search query
-Structure paper
| Title | Prokaryotic innate immunity through pattern recognition of conserved viral proteins. |
|---|---|
| Journal, issue, pages | Science, Vol. 377, Issue 6607, Page eabm4096, Year 2022 |
| Publish date | Aug 12, 2022 |
 Authors Authors | Linyi Alex Gao / Max E Wilkinson / Jonathan Strecker / Kira S Makarova / Rhiannon K Macrae / Eugene V Koonin / Feng Zhang /  |
| PubMed Abstract | Many organisms have evolved specialized immune pattern-recognition receptors, including nucleotide-binding oligomerization domain-like receptors (NLRs) of the STAND superfamily that are ubiquitous in ...Many organisms have evolved specialized immune pattern-recognition receptors, including nucleotide-binding oligomerization domain-like receptors (NLRs) of the STAND superfamily that are ubiquitous in plants, animals, and fungi. Although the roles of NLRs in eukaryotic immunity are well established, it is unknown whether prokaryotes use similar defense mechanisms. Here, we show that antiviral STAND (Avs) homologs in bacteria and archaea detect hallmark viral proteins, triggering Avs tetramerization and the activation of diverse N-terminal effector domains, including DNA endonucleases, to abrogate infection. Cryo-electron microscopy reveals that Avs sensor domains recognize conserved folds, active-site residues, and enzyme ligands, allowing a single Avs receptor to detect a wide variety of viruses. These findings extend the paradigm of pattern recognition of pathogen-specific proteins across all three domains of life. |
 External links External links |  Science / Science /  PubMed:35951700 / PubMed:35951700 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.9 - 3.48 Å |
| Structure data | EMDB-27421: Avs3 bound to phage PhiV-1 terminase, C2 refinement of Cap4 nuclease domain EMDB-27422: Avs4 bound to phage PhiV-1 portal, C2 refinement of Mrr nuclease domain 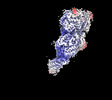 EMDB-27424: Avs3 bound to PhiV-1 terminase, symmetry-expanded C1 refinement of TPR-terminase domain 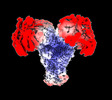 EMDB-27425: Avs4 bound to phage PhiV-1 portal, overall C2 reconstruction 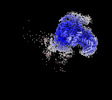 EMDB-27426: Avs4 bound to phage PhiV-1 portal, symmetry-expanded C1 refinement of TPR-portal domain |
| Chemicals |  ChemComp-ATP:  ChemComp-MG: |
| Source |
|
 Keywords Keywords |  ANTIVIRAL PROTEIN / phage defense / pattern-recognition receptor / nlr / stand / ANTIVIRAL PROTEIN / phage defense / pattern-recognition receptor / nlr / stand /  atpase atpase |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








 escherichia phage phiv-1 (virus)
escherichia phage phiv-1 (virus)