+Search query
-Structure paper
| Title | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 119, Issue 33, Page e2207200119, Year 2022 |
| Publish date | Aug 16, 2022 |
 Authors Authors | Jianhua Zhao / Suraj Makhija / Chenyu Zhou / Hanxiao Zhang / YongQiang Wang / Monita Muralidharan / Bo Huang / Yifan Cheng /  |
| PubMed Abstract | The ability to produce folded and functional proteins is a necessity for structural biology and many other biological sciences. This task is particularly challenging for numerous biomedically ...The ability to produce folded and functional proteins is a necessity for structural biology and many other biological sciences. This task is particularly challenging for numerous biomedically important targets in human cells, including membrane proteins and large macromolecular assemblies, hampering mechanistic studies and drug development efforts. Here we describe a method combining CRISPR-Cas gene editing and fluorescence-activated cell sorting to rapidly tag and purify endogenous proteins in HEK cells for structural characterization. We applied this approach to study the human proteasome from HEK cells and rapidly determined cryogenic electron microscopy structures of major proteasomal complexes, including a high-resolution structure of intact human PA28αβ-20S. Our structures reveal that PA28 with a subunit stoichiometry of 3α/4β engages tightly with the 20S proteasome. Addition of a hydrophilic peptide shows that polypeptides entering through PA28 are held in the antechamber of 20S prior to degradation in the proteolytic chamber. This study provides critical insights into an important proteasome complex and demonstrates key methodologies for the tagging of proteins from endogenous sources. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:35858375 / PubMed:35858375 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.7 - 3.4 Å |
| Structure data | EMDB-24275, PDB-7nan: EMDB-24276, PDB-7nao: EMDB-24277, PDB-7nap: EMDB-24278, PDB-7naq: EMDB-27013, PDB-8cvr:  EMDB-27014: Human PA28-20S proteasome with MG-132 (mixed PA28 subunits) EMDB-27015, PDB-8cvs:  EMDB-27016: Human 19S-20S proteasome, state SA  EMDB-27017: Human 19S-20S proteasome, state SD1 EMDB-27018, PDB-8cvt:  EMDB-27019: Human 19S-20S proteasome, state SD3/EC2 |
| Chemicals | 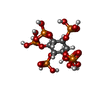 ChemComp-IHP:  ChemComp-LDZ:  ChemComp-ATP:  ChemComp-ZN: |
| Source |
|
 Keywords Keywords |  HYDROLASE / HYDROLASE /  proteasome / 20S / proteasome / 20S /  core particle / PA28 / PA200 / HYDROLASE/INHIBITOR / core particle / PA28 / PA200 / HYDROLASE/INHIBITOR /  proteolysis / proteolysis /  protein degradation / protein degradation /  complex / complex /  inhibitor / MG-132 / inhibitor / MG-132 /  MG132 / HYDROLASE-INHIBITOR complex MG132 / HYDROLASE-INHIBITOR complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



















