-Search query
-Search result
Showing all 30 items for (author: winter & sl)

EMDB-17993: 
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Method: single particle / : Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic J, Mathieu M, Bressanelli S

PDB-8pwh: 
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Method: single particle / : Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic J, Mathieu M, Bressanelli S

EMDB-18188: 
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Method: single particle / : Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic J, Mathieu M, Bressanelli S

PDB-8q6j: 
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Method: single particle / : Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic J, Mathieu M, Bressanelli S

EMDB-15244: 
Tomogram of an Ebola VLP composed of GP, VP40, NP, VP24 and VP35 at pH 7.4 (Figure 1A-D)
Method: electron tomography / : Winter SL, Chlanda P
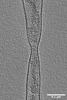
EMDB-15268: 
Tomogram of an Ebola VLP composed of VP40 at pH 4.5 (Figure 1J)
Method: electron tomography / : Winter SL, Chlanda P

EMDB-15951: 
Tomogram of an EBOV-infected Huh7 cell showing a late endosome with internalized EBOV particles
Method: electron tomography / : Winter SL, Chlanda P
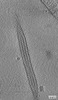
EMDB-15956: 
Tomogram of an extracellular EBOV particle adjacent to an EBOV-infected Huh7 cell
Method: electron tomography / : Winter SL, Chlanda P

EMDB-11791: 
MutS in Scanning state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D

EMDB-11792: 
MutS in mismatch bound state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D

EMDB-11793: 
MutS in Intermediate state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D

EMDB-11794: 
MutS-MutL in clamp state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D

EMDB-11795: 
MutS-MutL in clamp state (kinked clamp domain)
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D

PDB-7ai5: 
MutS in Scanning state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D, Sixma TK, Lamers MH

PDB-7ai6: 
MutS in mismatch bound state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D, Sixma TK, Lamers MH

PDB-7ai7: 
MutS in Intermediate state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D, Sixma TK, Lamers MH

PDB-7aib: 
MutS-MutL in clamp state
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D, Sixma TK, Lamers MH

PDB-7aic: 
MutS-MutL in clamp state (kinked clamp domain)
Method: single particle / : Fernandez-Leiro R, Bhairosing-Kok D, Sixma TK, Lamers MH

EMDB-10819: 
Tomogram of Lamellar body in A549 cell
Method: electron tomography / : Klein S, Wimmer B, Winter S, Kolovou A, Laketa V, Chlanda P

EMDB-10820: 
Tomogram of lamellar body (A549 cell expressing ABCA3-eGFP)
Method: electron tomography / : Klein S, Wimmer B, Winter S, Kolovou A, Laketa W, Chlanda P

EMDB-10818: 
Tomogram of Lamellar body in A549 cell expressing ABCA3-eGFP
Method: electron tomography / : Klein S, Wimmer B, Winter S, Kolovou A, Laketa V, Chlanda P

EMDB-11929: 
In situ cryo-ET of HSAEpC primary lung cells
Method: electron tomography / : Klein S, Wimmer B, Winter S, Kolovou A, Laketa V, Chlanda P

EMDB-11865: 
Tomogram of SARS-CoV-2 infected and cryo-FIB milled VeroE6 cells
Method: electron tomography / : Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P

EMDB-11931: 
Subtomogram average of outer membrane dome protein (OMDP)-C8 symmetrized
Method: subtomogram averaging / : Klein S, Wimmer B, Winter S, Kolovou A, Laketa V, Chlanda P

EMDB-11863: 
Cryo-ET of SARS-CoV-2 infected VeroE6 cells showing budding virions.
Method: electron tomography / : Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P

EMDB-11866: 
SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography
Method: electron tomography / : Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufleldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P

EMDB-11867: 
Cryo-ET of SARS-CoV-2 virions released from VerE6 cells
Method: electron tomography / : Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P

EMDB-11868: 
Subtomogram average of SARS-CoV-2 vRNP
Method: subtomogram averaging / : Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P

EMDB-21455: 
Cryo-EM structure of filamentous PFD from Methanocaldococcus jannaschii
Method: helical / : Wang F, Chen YX

PDB-6vy1: 
Cryo-EM structure of filamentous PFD from Methanocaldococcus jannaschii
Method: helical / : Wang F, Chen YX, Ing NL, Hochbaum AI, Clark DS, Glover DJ, Egelman EH
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
