[English] 日本語
 Yorodumi
Yorodumi- EMDB-15956: Tomogram of an extracellular EBOV particle adjacent to an EBOV-in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
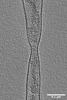
| Title | Tomogram of an extracellular EBOV particle adjacent to an EBOV-infected Huh7 cell | |||||||||
 Map data Map data | Tomogram of an extracellular EBOV particle adjacent to an EBOV-infected Huh7 cell | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Ebola virus / neutral pH / Ebola virus / neutral pH /  VIRUS VIRUS | |||||||||
| Biological species |   Ebola virus - Mayinga, Zaire, 1976 Ebola virus - Mayinga, Zaire, 1976 | |||||||||
| Method |  electron tomography / electron tomography /  cryo EM cryo EM | |||||||||
 Authors Authors | Winter SL / Chlanda P | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: The Ebola virus VP40 matrix layer undergoes endosomal disassembly essential for membrane fusion. Authors: Sophie L Winter / Gonen Golani / Fabio Lolicato / Melina Vallbracht / Keerthihan Thiyagarajah / Samy Sid Ahmed / Christian Lüchtenborg / Oliver T Fackler / Britta Brügger / Thomas Hoenen / ...Authors: Sophie L Winter / Gonen Golani / Fabio Lolicato / Melina Vallbracht / Keerthihan Thiyagarajah / Samy Sid Ahmed / Christian Lüchtenborg / Oliver T Fackler / Britta Brügger / Thomas Hoenen / Walter Nickel / Ulrich S Schwarz / Petr Chlanda /   Abstract: Ebola viruses (EBOVs) assemble into filamentous virions, whose shape and stability are determined by the matrix viral protein 40 (VP40). Virus entry into host cells occurs via membrane fusion in late ...Ebola viruses (EBOVs) assemble into filamentous virions, whose shape and stability are determined by the matrix viral protein 40 (VP40). Virus entry into host cells occurs via membrane fusion in late endosomes; however, the mechanism of how the remarkably long virions undergo uncoating, including virion disassembly and nucleocapsid release into the cytosol, remains unknown. Here, we investigate the structural architecture of EBOVs entering host cells and discover that the VP40 matrix disassembles prior to membrane fusion. We reveal that VP40 disassembly is caused by the weakening of VP40-lipid interactions driven by low endosomal pH that equilibrates passively across the viral envelope without a dedicated ion channel. We further show that viral membrane fusion depends on VP40 matrix integrity, and its disassembly reduces the energy barrier for fusion stalk formation. Thus, pH-driven structural remodeling of the VP40 matrix acts as a molecular switch coupling viral matrix uncoating to membrane fusion during EBOV entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15956.map.gz emd_15956.map.gz | 373.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15956-v30.xml emd-15956-v30.xml emd-15956.xml emd-15956.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15956.png emd_15956.png | 143.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15956 http://ftp.pdbj.org/pub/emdb/structures/EMD-15956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15956 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15956 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15956.map.gz / Format: CCP4 / Size: 507 MB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_15956.map.gz / Format: CCP4 / Size: 507 MB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomogram of an extracellular EBOV particle adjacent to an EBOV-infected Huh7 cell | ||||||||||||||||||||
| Voxel size | X=Y=Z: 8.013 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Ebola virus - Mayinga, Zaire, 1976
| Entire | Name:   Ebola virus - Mayinga, Zaire, 1976 Ebola virus - Mayinga, Zaire, 1976 |
|---|---|
| Components |
|
-Supramolecule #1: Ebola virus - Mayinga, Zaire, 1976
| Supramolecule | Name: Ebola virus - Mayinga, Zaire, 1976 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 128952 / Sci species name: Ebola virus - Mayinga, Zaire, 1976 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  electron tomography electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 / Focused ion beam - Current: 0.3 / Focused ion beam - Duration: 600 / Focused ion beam - Temperature: 100 K / Focused ion beam - Initial thickness: 6000 / Focused ion beam - Final thickness: 150 Focused ion beam - Details: The value given for _em_focused_ion_beam.instrument is Aquilos FIB-SEM. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 33000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 33000 |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number images used: 41 IMOD / Number images used: 41 |
|---|
 Movie
Movie Controller
Controller