-Search query
-Search result
Showing all 45 items for (author: zou & tt)

EMDB-17540: 
Cryo-EM structure of full-length human UBR5 (homotetramer)
Method: single particle / : Aguirre JD, Kater L, Kempf G, Cavadini S, Thoma NH

PDB-8p83: 
Cryo-EM structure of full-length human UBR5 (homotetramer)
Method: single particle / : Aguirre JD, Kater L, Kempf G, Cavadini S, Thoma NH

EMDB-17542: 
Negative stain map of UBR5 (dimer) in complex with RARA/RXRA
Method: single particle / : Aguirre JD, Cavadini S, Kempf G, Kater L, Thoma NH

EMDB-29714: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2
Method: single particle / : Ma MW, Hunkeler M, Jin CY, Fischer ES

PDB-8g46: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2
Method: single particle / : Ma MW, Hunkeler M, Jin CY, Fischer ES

EMDB-33226: 
Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom
Method: single particle / : Wang YJ, Guan ZY, Zou TT
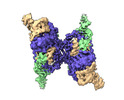
EMDB-31827: 
Cryo-EM structure of E.coli retron-Ec86 (RT-msDNA-RNA) at 3.2 angstrom
Method: single particle / : Wang YJ, Guan ZY, Zou TT
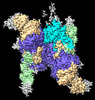
EMDB-31829: 
Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.8 angstrom
Method: single particle / : Wang YJ, Guan ZY, Zou TT

EMDB-13742: 
SARS-CoV-2 Spike ectodomain with Fab FI3A
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-7q0a: 
SARS-CoV-2 Spike ectodomain with Fab FI3A
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-12156: 
Structures of class II bacterial transcription complexes
Method: single particle / : Hao M, Ye FZ, Zhang XD

EMDB-12157: 
Structures of class I bacterial transcription complexes
Method: single particle / : Ye FZ, Hao M, Zhang XD

PDB-7bef: 
Structures of class II bacterial transcription complexes
Method: single particle / : Hao M, Ye FZ, Zhang XD

PDB-7beg: 
Structures of class I bacterial transcription complexes
Method: single particle / : Ye FZ, Hao M, Zhang XD

EMDB-10417: 
GroEL map obtained using the Preassis method for grid preparation
Method: single particle / : Zhao J, Xu H, Carroni M, Zou X

EMDB-10466: 
Cryo-ET on GroEL
Method: electron tomography / : Zou XD, Zhao JJ, Xu HY, Carroni M

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

PDB-7kdt: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-11173: 
Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab
Method: single particle / : Duyvesteyn HME, Zhou D, Zhao Y, Fry EE, Ren J, Stuart DI

PDB-6zdg: 
Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab
Method: single particle / : Duyvesteyn HME, Zhou D, Zhao Y, Fry EE, Ren J, Stuart DI

EMDB-10012: 
Apoferritin map obtained from grids prepared with the Preassis method
Method: single particle / : Zhao J, Xu H, Carroni M, Zou X

EMDB-11184: 
Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab
Method: single particle / : Duyvesteyn HME, Zhou D, Zhao Y, Fry EE, Ren J, Stuart DI

PDB-6zfo: 
Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab
Method: single particle / : Duyvesteyn HME, Zhou D, Zhao Y, Fry EE, Ren J, Stuart DI

EMDB-11174: 
SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab
Method: single particle / : Duyvesteyn HME, Zhou D, Zhao Y, Fry EE, Ren J, Stuart DI

PDB-6zdh: 
SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab
Method: single particle / : Duyvesteyn HME, Zhou D, Zhao Y, Fry EE, Ren J, Stuart DI

EMDB-4769: 
Cryo-EM structure of bacterial RNAP with a DNA mimic protein Ocr from T7 phage
Method: single particle / : Ye FZ, Zhang XD

EMDB-4770: 
Structural basis of transcription inhibition by the DNA mimic Ocr protein of bacteriophage T7
Method: single particle / : Ye FZ, Zhang XD

PDB-6r9b: 
Cryo-EM structure of bacterial RNAP with a DNA mimic protein Ocr from T7 phage
Method: single particle / : Ye FZ, Zhang XD

PDB-6r9g: 
Structural basis of transcription inhibition by the DNA mimic Ocr protein of bacteriophage T7
Method: single particle / : Ye FZ, Zhang XD

PDB-6qrz: 
Crystal structure of R2-like ligand-binding oxidase from Sulfolobus acidocaldarius solved by 3D micro-crystal electron diffraction
Method: electron crystallography / : Xu H, Lebrette H, Clabbers MTB, Zhao J, Griese JJ, Zou X, Hogbom M

EMDB-0199: 
human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA.
Method: single particle / : Oosterheert W, van Bezouwen LS, Rodenburg RNP, Forster F, Mattevi A, Gros P

EMDB-0200: 
human STEAP4 bound to NADPH, FAD and heme.
Method: single particle / : Oosterheert W, van Bezouwen LS, Rodenburg RNP, Forster F, Mattevi A, Gros P

PDB-6hcy: 
human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA.
Method: single particle / : Oosterheert W, van Bezouwen LS, Rodenburg RNP, Forster F, Mattevi A, Gros P

PDB-6hd1: 
human STEAP4 bound to NADPH, FAD and heme.
Method: single particle / : Oosterheert W, van Bezouwen LS, Rodenburg RNP, Forster F, Mattevi A, Gros P

EMDB-0001: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme transcription open complex
Method: single particle / : Glyde R, Ye FZ

EMDB-0002: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme intermediate partially loaded complex
Method: single particle / : Glyde R, Ye FZ

EMDB-4397: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex
Method: single particle / : Glyde R, Ye FZ

PDB-6gfw: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex
Method: single particle / : Glyde R, Ye FZ, Zhang XD

PDB-6gh5: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme transcription open complex
Method: single particle / : Glyde R, Ye FZ, Zhang XD

PDB-6gh6: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme intermediate partially loaded complex
Method: single particle / : Glyde R, Ye FZ, Zhang XD

PDB-5ocv: 
A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals
Method: electron crystallography / : Xu H, Lebrette H, Yang T, Srinivas V, Hovmoller S, Hogbom M, Zou X

EMDB-3506: 
cryoEM structure of bacterial holo-translocon
Method: single particle / : Schaffitzel C, Botte M, Karuppasamy M, Papai G, Schultz P

PDB-5mg3: 
EM fitted model of bacterial holo-translocon
Method: single particle / : Schaffitzel C, Botte M

EMDB-3282: 
Negative stain electron microscopy structure of TssA from EAEC type VI secretion system
Method: single particle / : Durand E, Fronzes R, Cambillau C, Cascales E
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
