+Search query
-Structure paper
| Title | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism. |
|---|---|
| Journal, issue, pages | Nat Microbiol, Vol. 7, Issue 9, Page 1480-1489, Year 2022 |
| Publish date | Aug 18, 2022 |
 Authors Authors | Yanjing Wang / Zeyuan Guan / Chen Wang / Yangfan Nie / Yibei Chen / Zhaoyang Qian / Yongqing Cui / Han Xu / Qiang Wang / Fen Zhao / Delin Zhang / Pan Tao / Ming Sun / Ping Yin / Shuangxia Jin / Shan Wu / Tingting Zou /  |
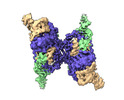
| PubMed Abstract | First discovered in the 1980s, retrons are bacterial genetic elements consisting of a reverse transcriptase and a non-coding RNA (ncRNA). Retrons mediate antiphage defence in bacteria but their ...First discovered in the 1980s, retrons are bacterial genetic elements consisting of a reverse transcriptase and a non-coding RNA (ncRNA). Retrons mediate antiphage defence in bacteria but their structure and defence mechanisms are unknown. Here, we investigate the Escherichia coli Ec86 retron and use cryo-electron microscopy to determine the structures of the Ec86 (3.1 Å) and cognate effector-bound Ec86 (2.5 Å) complexes. The Ec86 reverse transcriptase exhibits a characteristic right-hand-like fold consisting of finger, palm and thumb subdomains. Ec86 reverse transcriptase reverse-transcribes part of the ncRNA into satellite, multicopy single-stranded DNA (msDNA, a DNA-RNA hybrid) that we show wraps around the reverse transcriptase electropositive surface. In msDNA, both inverted repeats are present and the 3' sides of the DNA/RNA chains are close to the reverse transcriptase active site. The Ec86 effector adopts a two-lobe fold and directly binds reverse transcriptase and msDNA. These findings offer insights into the structure-function relationship of the retron-effector unit and provide a structural basis for the optimization of retron-based genome editing systems. |
 External links External links |  Nat Microbiol / Nat Microbiol /  PubMed:35982312 PubMed:35982312 |
| Methods | EM (single particle) |
| Resolution | 2.51 - 3.12 Å |
| Structure data | EMDB-31827, PDB-7v9u: EMDB-33226, PDB-7xjg: |
| Chemicals |  ChemComp-MG: |
| Source |
|
 Keywords Keywords | TRANSFERASE/DNA/RNA /  Reverse transcriptase / Reverse transcriptase /  RNA BINDING PROTEIN / TRANSFERASE-DNA-RNA complex / RNA BINDING PROTEIN/DNA/RNA / RNA BINDING PROTEIN-DNA-RNA complex RNA BINDING PROTEIN / TRANSFERASE-DNA-RNA complex / RNA BINDING PROTEIN/DNA/RNA / RNA BINDING PROTEIN-DNA-RNA complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers