-Search query
-Search result
Showing 1 - 50 of 124 items for (author: sine & s)

EMDB-42526: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine
Method: single particle / : Burke SM, Hibbs RE, Noviello CM

EMDB-42537: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and NS-1738
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-42841: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and ivermectin
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-43012: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and (-)-TQS
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-43015: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and PNU-120596
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-43025: 
Alpha7-nicotinic acetylcholine receptor bound to GAT107
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-43030: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and GAT107
Method: single particle / : Burke SM, Noviello CM, Hibbs RE
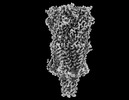
EMDB-43031: 
Alpha7-nicotinic acetylcholine receptor time resolved resting state
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-43032: 
Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 desensitized intermediate state
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-43034: 
Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-43035: 
Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 2
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8ut1: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine
Method: single particle / : Burke SM, Hibbs RE, Noviello CM

PDB-8utb: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and NS-1738
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8uzj: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and ivermectin
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v80: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and (-)-TQS
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v82: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and PNU-120596
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v86: 
Alpha7-nicotinic acetylcholine receptor bound to GAT107
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v88: 
Alpha7-nicotinic acetylcholine receptor bound to epibatidine and GAT107
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v89: 
Alpha7-nicotinic acetylcholine receptor time resolved resting state
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v8a: 
Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 desensitized intermediate state
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v8c: 
Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

PDB-8v8d: 
Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 2
Method: single particle / : Burke SM, Noviello CM, Hibbs RE

EMDB-40754: 
The 3alpha2beta stoichiometry of human alpha4beta2 nicotinic acetylcholine receptor in complex with acetylcholine and calcium
Method: single particle / : Kang G, Hibbs RE

EMDB-40755: 
The 3alpha2beta stoichiometry of human alpha4beta2 nicotinic acetylcholine receptor in complex with acetylcholine
Method: single particle / : Kang G, Hibbs RE

EMDB-40757: 
The 2alpha3beta stoichiometry of human alpha4beta2 nicotinic acetylcholine receptor in complex with acetylcholine
Method: single particle / : Kang G, Hibbs RE

EMDB-40756: 
The 2alpha3beta stoichiometry of human alpha4beta2 nicotinic acetylcholine receptor in complex with acetylcholine and calcium
Method: single particle / : Kang G, Hibbs RE

EMDB-40753: 
The 2alpha3beta stoichiometry of full-length human alpha4beta2 nicotinic acetylcholine receptor in complex with acetylcholine
Method: single particle / : Kang G, Hibbs RE

EMDB-40752: 
The 2alpha3beta stoichiometry of full-length human alpha4beta2 nicotinic acetylcholine receptor in complex with acetylcholine and calcium
Method: single particle / : Kang G, Hibbs RE

EMDB-40062: 
Kalium channelrhodopsin 1 from Hyphochytrium catenoides (HcKCR1) embedded in peptidisc
Method: single particle / : Morizumi T, Kim K, Li H, Spudich JL, Ernst OP

EMDB-40063: 
Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc
Method: single particle / : Morizumi T, Kim K, Li H, Spudich JL, Ernst OP

PDB-8gi8: 
Kalium channelrhodopsin 1 from Hyphochytrium catenoides (HcKCR1) embedded in peptidisc
Method: single particle / : Morizumi T, Kim K, Li H, Spudich JL, Ernst OP

PDB-8gi9: 
Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc
Method: single particle / : Morizumi T, Kim K, Li H, Spudich JL, Ernst OP

EMDB-29458: 
Alzheimer's disease paired-helical filament in complex with PET tracer GTP-1
Method: helical / : Merz GE, Tse E, Southworth DR

PDB-8fug: 
Alzheimer's disease paired-helical filament in complex with PET tracer GTP-1
Method: helical / : Merz GE, Tse E, Southworth DR

EMDB-28092: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28090: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-040
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO

EMDB-28091: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-045
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO
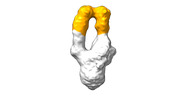
EMDB-28093: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156
Method: single particle / : Shek J, Callaway H, Li H, Yu X, Saphire EO
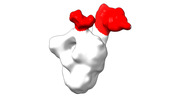
EMDB-28094: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-234
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28095: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-260
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28096: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-279
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28097: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-290
Method: single particle / : Yu X, Callaway H, Li H, Shek J, Saphire EO
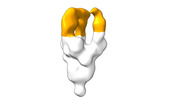
EMDB-28098: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-294
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28099: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-295
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28100: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-299
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28102: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-334
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO
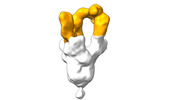
EMDB-28103: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-360
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28104: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-361
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28105: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-362
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28106: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-368
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
