+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1511 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | COPII coat | |||||||||
 Map data Map data | This is a reconstruction of a COPII coat comprised of Sec13-31 and Sec23-24. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  vesicle trafficking / vesicle trafficking /  COPII / icosidodecahedron / COPII / icosidodecahedron /  secretory pathway / secretory pathway /  endoplasmic reticulum / endoplasmic reticulum /  automation automation | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 43.0 Å cryo EM / Resolution: 43.0 Å | |||||||||
 Authors Authors | Stagg SM / LaPointe P / Razvi A / Gurkan C / Potter CS / Carragher B / Balch WE | |||||||||
 Citation Citation |  Journal: Cell / Year: 2008 Journal: Cell / Year: 2008Title: Structural basis for cargo regulation of COPII coat assembly. Authors: Scott M Stagg / Paul LaPointe / Abbas Razvi / Cemal Gürkan / Clinton S Potter / Bridget Carragher / William E Balch /  Abstract: Using cryo-electron microscopy, we have solved the structure of an icosidodecahedral COPII coat involved in cargo export from the endoplasmic reticulum (ER) coassembled from purified cargo adaptor ...Using cryo-electron microscopy, we have solved the structure of an icosidodecahedral COPII coat involved in cargo export from the endoplasmic reticulum (ER) coassembled from purified cargo adaptor Sec23-24 and Sec13-31 lattice-forming complexes. The coat structure shows a tetrameric assembly of the Sec23-24 adaptor layer that is well positioned beneath the vertices and edges of the Sec13-31 lattice. Fitting the known crystal structures of the COPII proteins into the density map reveals a flexible hinge region stemming from interactions between WD40 beta-propeller domains present in Sec13 and Sec31 at the vertices. The structure shows that the hinge region can direct geometric cage expansion to accommodate a wide range of bulky cargo, including procollagen and chylomicrons, that is sensitive to adaptor function in inherited disease. The COPII coat structure leads us to propose a mechanism by which cargo drives cage assembly and membrane curvature for budding from the ER. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1511.map.gz emd_1511.map.gz | 13.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1511-v30.xml emd-1511-v30.xml emd-1511.xml emd-1511.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1511.gif 1511.gif | 48.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1511 http://ftp.pdbj.org/pub/emdb/structures/EMD-1511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1511 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1511 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1511.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1511.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a reconstruction of a COPII coat comprised of Sec13-31 and Sec23-24. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
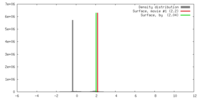
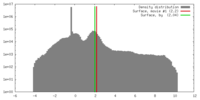
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Sec13/31 bound to Sec23/24
| Entire | Name: Sec13/31 bound to Sec23/24 |
|---|---|
| Components |
|
-Supramolecule #1000: Sec13/31 bound to Sec23/24
| Supramolecule | Name: Sec13/31 bound to Sec23/24 / type: sample / ID: 1000 Oligomeric state: 60 Sec13-31 heterotetramers bound to 120 Sec23-24 heterodimers Number unique components: 4 |
|---|---|
| Molecular weight | Theoretical: 44.8 MDa |
-Supramolecule #1: COPII coat
| Supramolecule | Name: COPII coat / type: organelle_or_cellular_component / ID: 1 / Name.synonym: Sec13-31 and Sec23-24 / Recombinant expression: Yes |
|---|---|
| Ref GO | divclassse qspanoncli ckpopupspa nclassgree n(this)spandata popltspanc lassquotlo adingbarqu otgtltimgs rcquotimgl oadinggifq uotdecodin gquotasync quotgtltsp angtdataur lajaxphp?m odetaxoamp ... divclassse qspanoncli ckpopupspa nclassgree n(this)spandata popltspanc lassquotlo adingbarqu otgtltimgs rcquotimgl oadinggifq uotdecodin gquotasync quotgtltsp angtdataur lajaxphp?m odetaxoamp kGO3A00068 88ampajax1 classpoptr giGO000688 8ispandiv |
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human / Location in cell: cytosol Homo sapiens (human) / synonym: Human / Location in cell: cytosol |
| Molecular weight | Experimental: 44.8 MDa / Theoretical: 44.8 MDa |
| Recombinant expression | Organism: Insect cells / Recombinant plasmid: bacmid |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 Details: 50 mM MES pH 6.8, 700 mM KOAc, 1 mM MgOAc, 1 mM DTT |
|---|---|
| Grid | Details: 400 mesh grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 98 K / Instrument: OTHER / Details: Vitrification instrument: vitrobot / Method: blot for 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 29000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus min: 10.0 µm / Nominal magnification: 29000 Bright-field microscopy / Cs: 2 mm / Nominal defocus min: 10.0 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Average: 88 K |
| Alignment procedure | Legacy - Astigmatism: astigmatism corrected automatically with Leginon. |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 20 e/Å2 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: each particle |
|---|---|
| Final two d classification | Number classes: 212 |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 43.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Details: Final map was lowpass filtered to 43 angstroms / Number images used: 12120 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 43.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Details: Final map was lowpass filtered to 43 angstroms / Number images used: 12120 |
| Details | Data was automatically collected using Leginon. Data was processed automatically with Appion. Particles were picked manually. |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: Normal mode-based flexible fitting (nmff) and chimera |
| Details | Protocol: normal modes flexible fitting and rigid body. A bend was modeled into Sec13-31 using the program nmff. Sec13-31 and Sec23-24 were separately fitted into the cryoEM density using the program Chimera. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name: nmff and chimera |
| Details | Protocol: normal modes flexible fitting and rigid body. A bend was modeled into Sec13-31 using the program nmff. Sec13-31 and Sec23-24 were separately fitted into the cryoEM density using the program Chimera. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
-Atomic model buiding 3
| Initial model | PDB ID: |
|---|---|
| Software | Name: nmff and chimera |
| Details | Protocol: normal modes flexible fitting and rigid body. A bend was modeled into Sec13-31 using the program nmff. Sec13-31 and Sec23-24 were separately fitted into the cryoEM density using the program Chimera. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)