+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4xnv | ||||||
|---|---|---|---|---|---|---|---|
| Title | The human P2Y1 receptor in complex with BPTU | ||||||
 Components Components | P2Y purinoceptor 1, Rubredoxin, P2Y purinoceptor 1 | ||||||
 Keywords Keywords | TRANSPORT PROTEIN/INHIBITOR / human P2Y1 receptor /  G protein coupled receptor / G protein coupled receptor /  platelet activation / platelet activation /  membrane protein / membrane protein /  inhibitor complex / TRANSPORT PROTEIN-INHIBITOR complex / PSI-Biology / inhibitor complex / TRANSPORT PROTEIN-INHIBITOR complex / PSI-Biology /  Structural Genomics / GPCR Network / Structural Genomics / GPCR Network /  GPCR GPCR | ||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled ATP receptor activity / relaxation of muscle /  A1 adenosine receptor binding / G protein-coupled ADP receptor activity / A1 adenosine receptor binding / G protein-coupled ADP receptor activity /  P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection ...G protein-coupled ATP receptor activity / relaxation of muscle / P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection ...G protein-coupled ATP receptor activity / relaxation of muscle /  A1 adenosine receptor binding / G protein-coupled ADP receptor activity / A1 adenosine receptor binding / G protein-coupled ADP receptor activity /  P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection / negative regulation of norepinephrine secretion / glial cell migration / regulation of presynaptic cytosolic calcium ion concentration / G protein-coupled adenosine receptor signaling pathway / positive regulation of hormone secretion / signal transduction involved in regulation of gene expression / response to growth factor / cellular response to ATP / regulation of synaptic vesicle exocytosis / P2Y receptors / cellular response to purine-containing compound / G protein-coupled purinergic nucleotide receptor activity / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of monoatomic ion transport / positive regulation of penile erection / negative regulation of norepinephrine secretion / glial cell migration / regulation of presynaptic cytosolic calcium ion concentration / G protein-coupled adenosine receptor signaling pathway / positive regulation of hormone secretion / signal transduction involved in regulation of gene expression / response to growth factor / cellular response to ATP / regulation of synaptic vesicle exocytosis /  eating behavior / monoatomic ion transport / presynaptic active zone membrane / response to mechanical stimulus / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / blood vessel diameter maintenance / protein localization to plasma membrane / establishment of localization in cell / eating behavior / monoatomic ion transport / presynaptic active zone membrane / response to mechanical stimulus / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / blood vessel diameter maintenance / protein localization to plasma membrane / establishment of localization in cell /  ADP binding / ADP binding /  cilium / cilium /  platelet activation / ADP signalling through P2Y purinoceptor 1 / platelet activation / ADP signalling through P2Y purinoceptor 1 /  signaling receptor activity / signaling receptor activity /  cell body / phospholipase C-activating G protein-coupled receptor signaling pathway / regulation of cell shape / positive regulation of cytosolic calcium ion concentration / cell body / phospholipase C-activating G protein-coupled receptor signaling pathway / regulation of cell shape / positive regulation of cytosolic calcium ion concentration /  scaffold protein binding / G alpha (q) signalling events / scaffold protein binding / G alpha (q) signalling events /  postsynaptic membrane / basolateral plasma membrane / postsynaptic membrane / basolateral plasma membrane /  postsynaptic density / postsynaptic density /  electron transfer activity / cell surface receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / positive regulation of protein phosphorylation / iron ion binding / apical plasma membrane / G protein-coupled receptor signaling pathway / protein heterodimerization activity / glutamatergic synapse / electron transfer activity / cell surface receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / positive regulation of protein phosphorylation / iron ion binding / apical plasma membrane / G protein-coupled receptor signaling pathway / protein heterodimerization activity / glutamatergic synapse /  dendrite / dendrite /  cell surface / positive regulation of transcription by RNA polymerase II / cell surface / positive regulation of transcription by RNA polymerase II /  ATP binding / ATP binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)  Clostridium pasteurianum (bacteria) Clostridium pasteurianum (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.2 Å SYNCHROTRON / Resolution: 2.2 Å | ||||||
 Authors Authors | Zhang, D. / Gao, Z. / Jacobson, K. / Han, G.W. / Stevens, R. / Zhao, Q. / Wu, B. / GPCR Network (GPCR) | ||||||
 Citation Citation |  Journal: Nature / Year: 2015 Journal: Nature / Year: 2015Title: Two disparate ligand-binding sites in the human P2Y1 receptor Authors: Zhang, D. / Gao, Z.G. / Zhang, K. / Kiselev, E. / Crane, S. / Wang, J. / Paoletta, S. / Yi, C. / Ma, L. / Zhang, W. / Han, G.W. / Liu, H. / Cherezov, V. / Katritch, V. / Jiang, H. / Stevens, ...Authors: Zhang, D. / Gao, Z.G. / Zhang, K. / Kiselev, E. / Crane, S. / Wang, J. / Paoletta, S. / Yi, C. / Ma, L. / Zhang, W. / Han, G.W. / Liu, H. / Cherezov, V. / Katritch, V. / Jiang, H. / Stevens, R.C. / Jacobson, K.A. / Zhao, Q. / Wu, B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4xnv.cif.gz 4xnv.cif.gz | 150.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4xnv.ent.gz pdb4xnv.ent.gz | 123.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4xnv.json.gz 4xnv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xn/4xnv https://data.pdbj.org/pub/pdb/validation_reports/xn/4xnv ftp://data.pdbj.org/pub/pdb/validation_reports/xn/4xnv ftp://data.pdbj.org/pub/pdb/validation_reports/xn/4xnv | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 47468.289 Da / Num. of mol.: 1 / Mutation: D320N Source method: isolated from a genetically manipulated source Details: Chimera protein of P2Y purinoceptor 1 (P2RY1_HUMA) with Rubredoxin (RUBR_CLOPA) inserted into ICL3 domain between residues 247LYS and 253PRO, and replaced residues 248ASN to 252SER. Source: (gene. exp.)   Homo sapiens (human), (gene. exp.) Homo sapiens (human), (gene. exp.)   Clostridium pasteurianum (bacteria) Clostridium pasteurianum (bacteria)Gene: P2RY1 / Production host:   Spodoptera (butterflies/moths) / References: UniProt: P47900, UniProt: P00268 Spodoptera (butterflies/moths) / References: UniProt: P47900, UniProt: P00268 |
|---|
-Non-polymers , 7 types, 45 molecules 











| #2: Chemical | ChemComp-BUR / | ||||||||
|---|---|---|---|---|---|---|---|---|---|
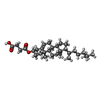
| #3: Chemical | ChemComp-CLR /  Cholesterol Cholesterol | ||||||||
| #4: Chemical | | #5: Chemical | ChemComp-OLC / ( #6: Chemical | ChemComp-1PE / |  Polyethylene glycol Polyethylene glycol#7: Chemical | ChemComp-ZN / | #8: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.11 Å3/Da / Density % sol: 41.8 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: lipidic cubic phase / Details: PEG2000MME Sodium Citrate |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 1 Å / Beamline: BL41XU / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jun 2, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→50 Å / Num. obs: 19881 / % possible obs: 99.9 % / Redundancy: 7.2 % / Biso Wilson estimate: 42.95 Å2 / Net I/σ(I): 12.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.2→41.4 Å / Cor.coef. Fo:Fc: 0.9412 / Cor.coef. Fo:Fc free: 0.9312 / SU R Cruickshank DPI: 0.293 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.265 / SU Rfree Blow DPI: 0.19 / SU Rfree Cruickshank DPI: 0.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 63.8 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.369 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→41.4 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.32 Å / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -11.1074 Å / Origin y: 17.9445 Å / Origin z: 9.7233 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: { A|39 - A|334 } |
 Movie
Movie Controller
Controller












 PDBj
PDBj





















