[English] 日本語
 Yorodumi
Yorodumi- PDB-4wmd: Crystal structure of catalytically inactive MERS-CoV 3CL protease... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4wmd | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of catalytically inactive MERS-CoV 3CL protease (C148A) in spacegroup C2221 | ||||||
 Components Components | ORF1a ORF1ab ORF1ab | ||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  3CL protease / MERS / 3CL protease / MERS /  coronavirus coronavirus | ||||||
| Function / homology |  Function and homology information Function and homology informationhost cell membrane / viral genome replication /  methyltransferase activity / methyltransferase activity /  methylation / symbiont-mediated degradation of host mRNA / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / methylation / symbiont-mediated degradation of host mRNA / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding /  omega peptidase activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / cysteine-type deubiquitinase activity ...host cell membrane / viral genome replication / omega peptidase activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / cysteine-type deubiquitinase activity ...host cell membrane / viral genome replication /  methyltransferase activity / methyltransferase activity /  methylation / symbiont-mediated degradation of host mRNA / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / methylation / symbiont-mediated degradation of host mRNA / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding /  omega peptidase activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / cysteine-type deubiquitinase activity / omega peptidase activity / symbiont-mediated perturbation of host ubiquitin-like protein modification / cysteine-type deubiquitinase activity /  single-stranded RNA binding / host cell perinuclear region of cytoplasm / single-stranded RNA binding / host cell perinuclear region of cytoplasm /  viral protein processing / virus-mediated perturbation of host defense response / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / induction by virus of host autophagy / cysteine-type endopeptidase activity / viral protein processing / virus-mediated perturbation of host defense response / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / induction by virus of host autophagy / cysteine-type endopeptidase activity /  proteolysis / zinc ion binding / proteolysis / zinc ion binding /  membrane membraneSimilarity search - Function | ||||||
| Biological species |   Middle East respiratory syndrome coronavirus Middle East respiratory syndrome coronavirus | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.585 Å MOLECULAR REPLACEMENT / Resolution: 2.585 Å | ||||||
 Authors Authors | Lountos, G.T. / Needle, D. / Waugh, D.S. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2015 Journal: Acta Crystallogr.,Sect.D / Year: 2015Title: Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Authors: Needle, D. / Lountos, G.T. / Waugh, D.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4wmd.cif.gz 4wmd.cif.gz | 188.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4wmd.ent.gz pdb4wmd.ent.gz | 150.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4wmd.json.gz 4wmd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wm/4wmd https://data.pdbj.org/pub/pdb/validation_reports/wm/4wmd ftp://data.pdbj.org/pub/pdb/validation_reports/wm/4wmd ftp://data.pdbj.org/pub/pdb/validation_reports/wm/4wmd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4wmeC  4wmfC  2ynaS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 3 molecules ABC
| #1: Protein |  ORF1ab ORF1abMass: 33322.109 Da / Num. of mol.: 3 / Mutation: C148A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Middle East respiratory syndrome coronavirus Middle East respiratory syndrome coronavirusPlasmid: pDN2551 / Production host:   Escherichia coli (E. coli) / References: UniProt: W6A941 Escherichia coli (E. coli) / References: UniProt: W6A941 |
|---|
-Non-polymers , 5 types, 291 molecules 
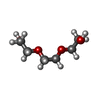
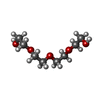






| #2: Chemical |  Diethylene glycol Diethylene glycol#3: Chemical |  Polyethylene glycol Polyethylene glycol#4: Chemical | ChemComp-PG4 / |  Polyethylene glycol Polyethylene glycol#5: Chemical | ChemComp-1PE / |  Polyethylene glycol Polyethylene glycol#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.28 Å3/Da / Density % sol: 71.3 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: 20.3 mg/mL protein, well solution: 0.1M Tris-Bis Tris pH 8.5, 0.12M ethylene glycols, 30% (v/v) ethylene glycol-polyethylene glycol 8000, (Morpheus Screeen condition E10). PH range: 8.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Aug 1, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.58→50 Å / Num. obs: 53763 / % possible obs: 99.9 % / Redundancy: 7.5 % / Rsym value: 0.077 / Net I/σ(I): 23.5 |
| Reflection shell | Resolution: 2.58→2.62 Å / Redundancy: 5.3 % / Rmerge(I) obs: 0.646 / Mean I/σ(I) obs: 2 / % possible all: 99.7 |
- Processing
Processing
| Software | Name: PHENIX / Version: (phenix.refine: dev_1448) / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: pdb entry 2YNA chain A Resolution: 2.585→46.162 Å / SU ML: 0.28 / Cross valid method: FREE R-VALUE / σ(F): 39.84 / Phase error: 23.01 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.585→46.162 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




