+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30416 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
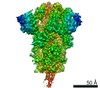
| Title | Cryo-EM structure of bat RaTG13 spike glycoprotein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationendocytosis involved in viral entry into host cell / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...endocytosis involved in viral entry into host cell / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Bat coronavirus RaTG13 Bat coronavirus RaTG13 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.93 Å cryo EM / Resolution: 2.93 Å | |||||||||
 Authors Authors | Wang X / Zhang S / Qiao S / Yu J / Zeng J / Tian L | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution. Authors: Shuyuan Zhang / Shuyuan Qiao / Jinfang Yu / Jianwei Zeng / Sisi Shan / Long Tian / Jun Lan / Linqi Zhang / Xinquan Wang /  Abstract: In recognizing the host cellular receptor and mediating fusion of virus and cell membranes, the spike (S) glycoprotein of coronaviruses is the most critical viral protein for cross-species ...In recognizing the host cellular receptor and mediating fusion of virus and cell membranes, the spike (S) glycoprotein of coronaviruses is the most critical viral protein for cross-species transmission and infection. Here we determined the cryo-EM structures of the spikes from bat (RaTG13) and pangolin (PCoV_GX) coronaviruses, which are closely related to SARS-CoV-2. All three receptor-binding domains (RBDs) of these two spike trimers are in the "down" conformation, indicating they are more prone to adopt the receptor-binding inactive state. However, we found that the PCoV_GX, but not the RaTG13, spike is comparable to the SARS-CoV-2 spike in binding the human ACE2 receptor and supporting pseudovirus cell entry. We further identified critical residues in the RBD underlying different activities of the RaTG13 and PCoV_GX/SARS-CoV-2 spikes. These results collectively indicate that tight RBD-ACE2 binding and efficient RBD conformational sampling are required for the evolution of SARS-CoV-2 to gain highly efficient infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30416.map.gz emd_30416.map.gz | 8.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30416-v30.xml emd-30416-v30.xml emd-30416.xml emd-30416.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30416_fsc.xml emd_30416_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_30416.png emd_30416.png | 75.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30416 http://ftp.pdbj.org/pub/emdb/structures/EMD-30416 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30416 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30416 | HTTPS FTP |
-Related structure data
| Related structure data |  7cn4MC  7cn8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30416.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30416.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RaTG13 glycoprotein
| Entire | Name: RaTG13 glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: RaTG13 glycoprotein
| Supramolecule | Name: RaTG13 glycoprotein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Bat coronavirus RaTG13 Bat coronavirus RaTG13 |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bat coronavirus RaTG13 Bat coronavirus RaTG13 |
| Molecular weight | Theoretical: 140.512719 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SSTRGVYYPD KVFRSSVLHL TQDLFLPFFS NVTWFHAIHV SGTNGIKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SSTRGVYYPD KVFRSSVLHL TQDLFLPFFS NVTWFHAIHV SGTNGIKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPP GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTDSI VRFPNITNLC PFGEVFNATT FASVYAWNRK RISNCVADYS VLYNSTSFST FKCYGVSPTK LNDLCFTNVY ADSF VITGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSKHIDAKE GGNFNYLYRL FRKANLKPFE RDISTEIYQA GSKPC NGQT GLNCYYPLYR YGFYPTDGVG HQPYRVVVLS FELLNAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN ASNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSRS VASQSIIAYT MSLGAENSVA YSNNSIAIPT NFTISVTT E ILPVSMTKTS VDCTMYICGD STECSNLLLQ YGSFCTQLNR ALTGIAVEQD KNTQEVFAQV KQIYKTPPIK DFGGFNFSQ ILPDPSKPSK RSFIEDLLFN KVTLADAGFI KQYGDCLGDI AARDLICAQK FNGLTVLPPL LTDEMIAQYT SALLAGTITS GWTFGAGAA LQIPFAMQMA YRFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLSSTAS ALGKLQDVVN QNAQALNTLV K QLSSNFGA ISSVLNDILS RLDPPEAEVQ IDRLITGRLQ SLQTYVTQQL IRAAEIRASA NLAATKMSEC VLGQSKRVDF CG KGYHLMS FPQSAPHGVV FLHVTYVPAQ EKNFTTAPAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSG SCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELG KYEQY IKWPLEVLFQ GPGGGSGGGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGHH HHHHWSHPQF EK |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 36 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller